Abstract
Objectives
To investigate if quantitative apparent diffusion coefficient (ADC) measurements can predict genetic subtypes of non-gadolinium-enhancing gliomas, comparing whole tumour against single slice analysis.
Methods
Volumetric T2-derived masks of 44 gliomas were co-registered to ADC maps with ADC mean (ADCmean) calculated. For the slice analysis, two observers placed regions of interest in the largest tumour cross-section. The ratio (ADCratio) between ADCmean in the tumour and normal appearing white matter was calculated for both methods.
Results
Isocitrate dehydrogenase (IDH) wild-type gliomas showed the lowest ADC values throughout (p < 0.001). ADCmean in the IDH-mutant 1p19q intact group was significantly higher than in the IDH-mutant 1p19q co-deleted group (p < 0.01). A volumetric ADCmean threshold of 1201 × 10−6 mm2/s identified IDH wild-type with a sensitivity of 83% and a specificity of 86%; a volumetric ADCratio cut-off value of 1.65 provided a sensitivity of 80% and a specificity of 92% (area under the curve (AUC) 0.9–0.94). A slice ADCratio threshold for observer 1 (observer 2) of 1.76 (1.83) provided a sensitivity of 80% (86%), specificity of 91% (100%) and AUC of 0.95 (0.96). The intraclass correlation coefficient was excellent (0.98).
Conclusions
ADC measurements can support the distinction of glioma subtypes. Volumetric and two-dimensional measurements yielded similar results in this study.
Key Points
• Diffusion-weighted MRI aids the identification of non-gadolinium-enhancing malignant gliomas
• ADC measurements may permit non-gadolinium-enhancing glioma molecular subtyping
• IDH wild-type gliomas have lower ADC values than IDH-mutant tumours
• Single cross-section and volumetric ADC measurements yielded comparable results in this study
Similar content being viewed by others
Introduction
Gadolinium contrast uptake was previously considered the best MR imaging predictor of glioma histological grade and malignancy[1,2,3]. On the basis of this, it has been common practice to interpret non-enhancing intrinsic tumours as probable low grade gliomas (LGG) [4]. But conventional MRI has proven to be unreliable in predicting subsequent tumour behaviour, whereby a proportion of presumed LGG may rapidly progress with development of malignant features such as enhancement and necrosis [4,5,6,7,8].
The discovery of several key genetic alterations as principal determinants of glioma prognosis has challenged the reference standard of glioma grouping by histology [9]. Mutations in isocitrate dehydrogenase (IDH) represent a common (> 70%) defining event in the development of LGG, conversely more than 90% of glioblastomas belong to the IDH wild-type group [10, 11]. Despite its oncogenic effect through production of a toxic metabolite D2-hydroxyglutarate (2HG), the presence of an IDH mutation is associated with a favourable prognosis.
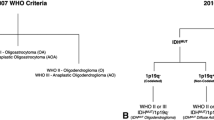
The revised 2016 World Health Organization (WHO) classification of brain tumours for the first time incorporates molecular data to augment the diagnosis [12]. For WHO grade II/III gliomas, three molecular subgroups have been defined: IDH wild-type glioma (IDHwt) with survival similar to that of glioblastoma, IDH-mutant glioma with intact 1p19q (IDHmut1p19int) and an intermediate prognosis, and IDH-mutant 1p19q co-deleted glioma (IDHmut1p19qdel) with the best prognosis and greatest chemosensitivity [11]. There is partial overlap with histomorphology, whereby many IDHmut1p19int are astrocytic and the majority of IDHmut1p19qdel belong to the oligodendroglioma group [13]. IDHwt gliomas probably constitute a genetically heterogeneous category of lesions, but often exhibit aggressive behaviour and have been suspected to represent early glioblastoma [14,15,16,17]. In the emerging literature on MR imaging features of IDHwt glioma, initial lack of enhancement has been reported in some of these tumours [6, 18, 19].
Diffusion-weighted imaging (DWI) is a technique of great interest in cancer, because water diffusivity is impaired in highly cellular tissues, which reflects tumour proliferative rate and aggressiveness [20]. The phenomenon of reduced diffusion preceding fulminant radiological progression of presumed LGG has been observed prior to molecular typing [7], evoking later descriptions of IDHwt glioma serial imaging findings [4]. Quantitative apparent diffusion coefficient (ADC) values have demonstrated high accuracy for glioma grading through meta-analysis [21]. For the non-invasive identification of low to intermediate IDHwt glioma, diffusion tensor imaging (DTI) and diffusion kurtosis imaging (DKI) have shown potential, suggesting that reduced and heterogenous diffusivity are IDHwt features [22,23,24]. However, advanced diffusion techniques are not universally available outside academic hospital institutions, may require longer scan times and dedicated post-processing.
Mean ADC measurement could be a rapid and practicable approach to assess glioma diffusivity, being computationally non-demanding compared to histograms or texture analysis. Although theoretically superior, there is no conclusive evidence that whole lesion analysis outperforms region-of-interest placement for the identification of malignant gliomas [25].
The study presented sought to (i) investigate whether ADC measurements from routine clinical DWI were associated with glioma molecular subtype and (ii) to compare the performance of volumetric whole tumour ADC with single slice ADC measurements.
Materials and methods
Patients
Following institutional board approval for a retrospective study, we searched the neuropathology records revealing 37 patients with WHO grade II/III IDHwt glioma between 2009 and 2016. For comparison of the molecular groups, control samples of IDH (IDH1-R132H) mutant gliomas (34 IDHmut1p19qint and 32 IDHmut1p19qdel) were randomly selected. We sought to evaluate ADC for suspected LGG prior to tissue diagnosis. To replicate the clinical situation, only gliomas without gadolinium enhancement were included (2 non-enhancing gliomas were excluded because of missing images and degraded DWI, respectively). The study sample consisted of 14 IDHwt (7 WHO II and 7 WHO III), 16 IDHmut1p19qint (8 WHO II and 8 WHO III) and 14 IDHmut1p19qdel (11 WHO II and 3 WHO III), amounting to 44 non-enhancing gliomas for the three molecular groups (patient selection diagram shown in Fig. 1). No haemorrhagic or necrotic gliomas were featured in the study.
MRI acquisition
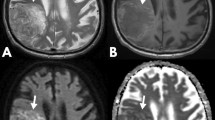
Ours is a quaternary neurosurgical centre; therefore the standard (structural and DWI) MRI sequences in this study originated from 10 different referring institutions (institution 1 to institution 10): 29 from our own institutions, 4 from institution 2, 3 from institution 3, 2 from institution 4, and one each from the remaining six institutions. The studies were acquired on 18 different scanners (31 at 1.5 Tesla, and 13 at 3 Tesla) from all major vendors: four General Electric scanners [Discovery MR450 (number of patients n = 5), 2× Signa Excite (n = 1 each), Genesis Signa (n = 2)], seven Siemens scanners [3× Avanto (n = 7, n = 2, n = 1), a Trio (n = 9), Symphony (n = 4), Skyra (n = 3), Espree (n = 1)], six Philips scanners [Ingenia (n = 2), 5× Achieva (n = 1 each)] and one Toshiba scanner (n = 1). All acquisitions included axial T2-weighted images, and axial standard 3-directional whole brain DWI. The median [min, max] values of the parameters of the T2-weighted images were echo time (TE) = 99.5 [80, 141] ms; repetition time (TR) = 4610 [2500, 7480] ms, in-plane resolution = 0.5 × 0.5 [0.3 × 0.3, 0.9 × 0.9] mm2; slice thickness = 5 [1, 6] mm; gap between slices = 1.5 [0, 2] mm. All DWI acquisitions included diffusion gradient weighting values b = 0 s/mm2 and b = 1000 s/mm2; the median [min, max] of other parameters were TE = 90.5 [69.5, 137] ms; TR = 4000 [2837, 10,000] ms, in-plane resolution = 1.25 × 1.25 [0.5 × 0.5, 2.5 × 2.5] mm2; slice thickness = 5 [4, 6] mm; gap between slices = 1.5 [0, 2] mm. For each patient, the imaging study was performed on average (standard deviation, sd) 2.3 (2.8) months prior to the tissue diagnosis. Image examples for the glioma molecular subgroups are shown in Fig. 2.
Post-processing and ADC analysis
ADC map calculation
In a spin echo diffusion-weighted sequence, the signal Sb [Sb = S0 e(−b ADC)] from each pixel in an image is formed of a first component (S0) dependent on tissue properties (i.e. ‘spin density’, T1 and T2 relaxation times) and sequence properties (e.g. repetition time, TR); and a second component (e−b ADC) dependent on the diffusion gradients (b, in units of s/mm2) and the apparent diffusion coefficient (ADC, in units of mm2/s).
The ADC is obtained by dividing the image acquired without diffusion gradients (Sb = 0 = S0) by the image acquired with diffusion gradients (Sb):
In this division, the dependence of ADC from S0 (and therefore from T1, T2 and TR) is eliminated [26]. The ADC maps were calculated using Eq. 1 and the utility fslmaths from the software library fsl (version 5.0) [27]. Offline whole tumour analysis and single slice analysis were subsequently performed.
Whole tumour (volumetric) ADC analysis
Tumour volumes of interest (VOItum) were outlined by a neuroradiology resident (S.H.) using ITK snap Toolbox version 3.6 (www.itksnap.org [28]), covering the entire T2 signal abnormality with each segmentation optimised by a board-certified neuroradiologist specialised in brain tumour imaging (S.C.T.). For multicentric gliomas, the total volume of signal abnormality was treated as one lesion. ADC maps were co-registered to T2 imaging using the FLIRT toolbox [29, 30] performing a rigid body transformation with a six-parameter model and ‘Normalised Mutual Information’ as cost function. Subsequently, ADCmean measurements were obtained for each tumour, using the fslstats utility from fsl [25,26,27].
To consider possible interindividual variations in brain diffusivity, we assessed the ADCmean in normal appearing white matter (NAWM). For each patient, a standardised second volume of interest (VOICS) was drawn in the contralateral centrum semiovale (CS). This VOICS was used to calculate the ADCratio = ADCmean(VOItum)/ADCmean(VOICS) (Fig. 2). For two IDHwt tumours, the NAWM analysis was omitted because of bilateral tumour infiltration.
Single slice ADC analysis
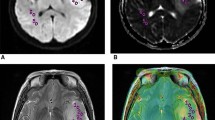
Standard picture archiving and communication systems (PACS) software (IMPAX 6.5.1.1008, Agfa-Gevaert, Mortsel, Belgium) was used to exploit tools routinely available for reporting of MR images. Two observers blinded to histomolecular results (J.A.M. general radiology trainee = observer 1 and S.C.T. = observer 2) located the tumour on the T2-weighted sequence, selecting two round regions of interest on the ADC map viewed side-by-side: The first region of interest (ROItum) was drawn in the largest lesion cross-section sparing the tumour margin to avoid partial volume effects. The second round ROICS aiming for a similar size to ROItum was placed in contralateral centrum semiovale NAWM, taking care to exclude images with visible ventricular surfaces, cortex and/or sulcal spaces at measurement level. Three patients were excluded from the single slice analysis because of non-availability of an ADC map on PACS. The ratio between the ADCmean in the tumour and CS was calculated [PACS_ADCratio = ADCmean(ROItum)/ADCmean(ROICS)]. No absolute ADC values were measured by the single slice method, as their workstation display can vary depending on the referring institution. An example of the volumetric segmentation and single slice ADC measurement is demonstrated in Fig. 3.
Histopathology and molecular analysis
Paraffin blocks containing tissue were analysed at our institution’s neuropathology department according to WHO 2016 guidance and previously published data [16]. IDH R132H immuno-negative tumours underwent multiple gene Sanger sequencing. A quantitative polymerase chain reaction-based copy number assay was used to determine 1p/19q status.
Statistical analysis
For the volumetric and single slice data, the statistical analysis consisted of two steps each: (i) linear regression to assess the association between the tumour type (IDHwt, IDHmut1p19qint, IDHmut1p19qdel) and ADC values, followed by (ii) logistic regression to determine if ADC values can differentiate IDHwt from IDHmut gliomas. A receiver operating characteristic (ROC) analysis was used to quantify the performance of the logistic regression. For the identification of a cut-off point for the logistic regression the ‘nearest to (0,1)’ method was performed. Statistical significance was set at 5%. The inter-rater agreement was expressed as an intraclass correlation coefficient (ICC) using a two-way random effects model. All statistical analyses were performed using Stata version 14 (College Station, TX: StataCorp LP).
Results
The mean age was greater in the IDHwt group than in the IDHmut groups (p = 0.0001 for IDHmut1p19qint, p = 0.005 for IDHmut1p19qdel). The larger proportion of WHO II gliomas in the IDHmut1p19qdel was not statistically significant (Pearson chi-square test p = 0.115 for IDHwt and p = 0.105 for IDHmut1p19qint). The patient demographic data and tumour volumes are reported in Table 1.
Association between molecular subtype and ADC values
In the volumetric analysis, IDHwt tumours showed significantly lower whole tumour volume ADCmean(VOItum) than IDHmut1p19qint (p < 0.0005) and IDHmut1p19qdel (p = 0.001). The ADCmean(VOItum) in the IDHmut1p19qint group was significantly higher than in the IDHmut1p19qdel group (p = 0.0047).
IDHwt gliomas had a significantly lower whole tumour ADCratio than IDHmut1p19qint (p < 0.0005) and IDHmut1p19qdel (p = 0.019). The ADCratio in the IDHmut1p19qint group was significantly higher than in the IDHmut1p19qdel group (p = 0.0054).
On single slice assessment, a significantly lower mean PACS_ADCratio was observed for IDHwt than for IDHmut1p19qint (p < 0.0005 observer 1; p < 0.0005 observer 2) and for IDHmut1p19qdel (p = 0.001 observer 1; p = 0.001 observer 2). The PACS_ADCratio in the IDHmut1p19qint group was higher than in the IDHmut1p19qdel group (p = 0.0008 for observer 1 and p = 0.0025 for observer 2). No statistical associations were demonstrated between the NAWM ADCmean values and molecular subtype.
The intra-rater agreement for the PACS_ADCratio measurements was very high: the correlation of measurements made on the same individual was 0.96, while the correlation between mean observer ratings was 0.98. The correlation of measurements equaled the consistency agreement, indicating no systematic difference between the two observers. The single slice ADCratio values were slightly but systematically higher than the volumetric ADCratio. The numerical results of the association between tumour type and ADC values for the volumetric and single slice analyses are reported in Table 2. In Table 3, the difference between the ADC values in IDHmut1p19qint and in IDHmut1p19qdel is shown. In Table 4 the ICC values are detailed. The boxplots of the ADCmean and ADCratio values are depicted in Fig. 4.
Diagnostic performance of ADC values
For ADCmean(VOItum), a ROC analysis quantified the accuracy of correctly classifying tumour type to an area under the curve (AUC) of 0.94. The cut-off point for the ADCmean(VOItum) was 1201 × 10−6 mm2/s, with a sensitivity of 0.83 and a specificity of 0.86. For a decrease in the ADCmean(VOItum) value by 1.0 × 10−5 mm2/s, the odds of IDHwt increased by 78% (p = 0.003).
For the volumetric ADCratio, the ROC analysis yielded an AUC of 0.90 with a sensitivity of 0.80 and a specificity of 0.92 for a threshold ADCratio of 1.65. For a decrease in the volumetric ADCratio value by 0.1, the odds of IDHwt increased by 46% (p = 0.004).
A ROC analysis quantified the accuracy of the PACS_ADCratio logistic regression in correctly classifying tumour type to an AUC of 0.96 for observer 1 and 0.95 for observer 2. The cut-off point for the PACS_ADCratio for observer 1 (observer 2) was 1.83 (1.76) with a sensitivity of 0.80 (0.86) and a specificity of 1.00 (0.91) at the cut-off point. For a decrease in the single slice ADCratio value by 0.1, the odds of IDHwt increased by 62% (p = 0.005) for observer 1 and 57% (p = 0.004) for observer 2. The numerical results for glioma subtype prediction are reported in Table 5. The ROC curves are depicted in Fig. 5.
Discussion
In this analysis, we observed that ADC values obtained from standard clinical DWI are a highly significant predictor of non-enhancing glioma IDH status and may permit non-invasive molecular subtyping in accordance with the 2016 WHO classification.
Two recent surveys highlighted clinical practices in caring for patients with presumed LGG, with approximately 50% of neurosurgeons adopting a ‘wait and see’ approach balanced against surgical risk [31], and only 21% performing an upfront biopsy [32]. Consequently, innocuous appearing IDHwt gliomas may reveal their aggressive nature through progression and receive treatment with a delay.
Low ADC values are associated with increased glioma cellularity and worse prognosis, supported by comparisons of diffusivity, histological specimens and clinical data in multiple studies [5, 33,34,35,36,37]. Low diffusivity predicts poor astrocytoma survival independent from WHO grade [38], although no linear relation exists between ADC and glioma prognosis [39].
Past studies to distinguish astrocytoma and oligodendroglioma using ADC values yielded variable success [40, 41], and in retrospect may have been influenced by the incomplete overlap between histological and molecular groups. Diagnostic focus has shifted to genetic typing, yet immunohistochemistry tests are complex and not infallible, requiring interpretation in in the context of morphological criteria and test type performed to avoid interpretational errors [42].
Recently, Leu et al. were able to assign gliomas to the WHO 2016 molecular groups using ADC; however, their method differed from ours by including enhancing lesions and ADC median values derived from b700–1000 gradients with DTI analysed for some patients [43]. To our best knowledge, this is the first IDH typing study to focus on non-enhancing gliomas, using b1000 values derived from 3-directional DWI. This is particularly important, as such tumours are usually assumed to be less aggressive in common clinical practice.
We found ADCratio values to be closely reproducible when comparing whole lesion measurements against single slice region of interest placements, for which there was near complete interobserver agreement. The similarity of our volumetric and single slice results could be explained by a relative homogeneity of these non-enhancing, non-necrotic gliomas. Both the absolute ADCmean values and ADCratio appear valuable for this lesion type. The quicker and easier single slice analysis even performed marginally better. This is in line with results of previous imaging research, which suggested that whole lesion diffusivity measurement is not always superior to ROI analysis [25, 44].
The ability of ADC to predict glioma subtypes and optimum thresholds may be subject to ROI placement technique with previous research focusing on minimum ADC value analysis: Xing et al. showed a statistical correlation between ADC and IDH status using a multiple (≥ 5) ROI technique with the mean of the lowest ADC measurement chosen as minimum ADC in consensus [45]. In a similar fashion, a previous DTI study for IDH typing used multiple ROI placements and a two-reader consensus method to obtain minimum ADC values [24].
As a reference ROI, we chose the centrum semiovale for its potentially greater reproducibility compared to a ‘mirror’ ROI [45], because this could be influenced by tumour location. We avoided the internal capsule [24], which is a smaller structure and more difficult to locate by an untrained rater.
Lee et al. found ADC mean and ADC histograms useful for IDH typing of WHO grade III and IV gliomas [46]. However, for glioblastoma IDH typing alone, a recent study identified no difference in ADC values [47]. In Tan et al.’s study of grade II–IV gliomas, the accuracy of ADC for IDH typing decreased with higher grade, which may reflect greater lesion heterogeneity [24]. It is probable that in such circumstances advanced diffusion acquisitions (e.g. DKI or multi-b-value imaging) could provide greater tissue microstructural information.
The good performance of the single slice ROI technique in IDH typing of non-enhancing lower grade gliomas was unexpected, but is highly relevant. It implies that such easy-to-perform measurements could be incorporated into clinical reports, complementing advanced MR modalities such as perfusion and 2HG spectroscopy [48, 49] pending tissue diagnosis. The origin of data from 18 MRI systems could represent a limitation of this study, but reflects clinical reality. The fact that significant separation of glioma subtypes could be obtained from this dataset further underscores the robustness of ADC.
It remains unknown why intermediate ADC values were observed in the 1p19q co-deleted gliomas, despite their best prognosis. This result is consistent with published data on intermediate diffusivity in oligodendroglioma; interestingly this tumour subtype may also mimic malignant gliomas on MR perfusion studies [39, 50].
In summary, the results from this study suggest that for newly diagnosed non-enhancing gliomas with ADC ratio values of 1.8 or less, further investigation with consideration of early tissue diagnosis is advisable given an increased risk of IDHwt molecular status.
Conclusions
ADC measurement appears to be a simple and powerful method for molecular subtyping of non-enhancing WHO II–III gliomas, specifically to identify IDHwt neoplasms. In our patient cohort, a two-dimensional ROI measurement in the largest lesion cross-section appeared representative of the entire tumour with comparable results.
Abbreviations
- ADC:
-
Apparent diffusion coefficient
- AUC :
-
Area under the curve
- CS:
-
Centrum semiovale
- DKI:
-
Diffusion kurtosis imaging
- DTI:
-
Diffusion tensor imaging
- DWI:
-
Diffusion-weighted imaging
- IDH :
-
Isocitrate dehydrogenase
- IDHwt :
-
Isocitrate dehydrogenase wild-type
- IDHmut1p19int :
-
Isocitrate dehydrogenase-mutant 1p19q intact
- IDHmut1p19del :
-
Isocitrate dehydrogenase-mutant 1p19q co-deleted
- ICC:
-
Intraclass correlation coefficient
- LGG:
-
Low grade glioma
- NAWM:
-
Normal appearing white matter
- PACS:
-
Picture archiving and communications system
- ROC:
-
Receiver operating characteristic
- ROICS :
-
Centrum semiovale region of interest
- ROItum :
-
Tumour region of interest
- TE:
-
Echo time
- TR :
-
Repetition time
- VOICS :
-
Centrum semiovale volume of interest
- VOItum :
-
Tumour volume of interest
- WHO :
-
World Health Organization
- 2HG :
-
D2-hydroxyglutarate
References
Pierallini A, Bonamini M, Bozzao A et al (1997) Supratentorial diffuse astrocytic tumours: proposal of an MRI classification. Eur Radiol 7:395–399
Watanabe M, Tanaka R, Takeda N (1992) Magnetic resonance imaging and histopathology of cerebral gliomas. Neuroradiology 34:463–469
Tervonen O, Forbes G, Scheithauer BW, Dietz MJ (1992) Diffuse “fibrillary” astrocytomas: correlation of MRI features with histopathologic parameters and tumor grade. Neuroradiology 34:173–178
Ideguchi M, Kajiwara K, Goto H et al (2015) MRI findings and pathological features in early-stage glioblastoma. J Neurooncol 123:289–297
Cuccarini V, Erbetta A, Farinotti M et al (2016) Advanced MRI may complement histological diagnosis of lower grade gliomas and help in predicting survival. J Neurooncol 126:279–288
Olar A, Raghunathan A, Albarracin CT et al (2012) Absence of IDH1-R132H mutation predicts rapid progression of nonenhancing diffuse glioma in older adults. Ann Diagn Pathol 16:161–170
Baehring JM, Bi WL, Bannykh S et al (2007) Diffusion MRI in the early diagnosis of malignant glioma. J Neurooncol 82:221–225
Ginsberg LE, Fuller GN, Hashmi M et al (1998) The significance of lack of MR contrast enhancement of supratentorial brain tumors in adults: histopathological evaluation of a series. Surg Neurol 49:436–440
Dang L, White DW, Gross S et al (2009) Cancer-associated IDH1 mutations produce 2-hydroxyglutarate. Nature 462:739–744
Yan H, Parsons DW, Jin G et al (2009) IDH1 and IDH2 mutations in gliomas. N Engl J Med 360:765–773
Cancer Genome Atlas Research Network, Brat DJ, RGW V et al (2015) Comprehensive, integrative genomic analysis of diffuse lower-grade gliomas. N Engl J Med 372:2481–2498
Louis DN, Perry A, Burger P et al (2014) International Society of Neuropathology–Haarlem consensus guidelines for nervous system tumor classification and grading. Brain Pathol Zurich Switz 24:429–435
Eckel-Passow JE, Lachance DH, Molinaro AM et al (2015) Glioma groups based on 1p/19q, IDH, and TERT promoter mutations in tumors. N Engl J Med 372:2499–2508
Abudumijiti A, Chan AK-Y, Shi Z et al (2017) Adult IDH wild-type lower-grade gliomas should be further stratified. Neuro Oncol. https://doi.org/10.1093/neuonc/nox078
Suzuki Y, Shirai K, Oka K et al (2010) Higher pAkt expression predicts a significant worse prognosis in glioblastomas. J Radiat Res (Tokyo) 51:343–348
Reuss DE, Sahm F, Schrimpf D et al (2015) ATRX and IDH1-R132H immunohistochemistry with subsequent copy number analysis and IDH sequencing as a basis for an “integrated” diagnostic approach for adult astrocytoma, oligodendroglioma and glioblastoma. Acta Neuropathol (Berl) 129:133–146
Hartmann C, Hentschel B, Wick W et al (2010) Patients with IDH1 wild type anaplastic astrocytomas exhibit worse prognosis than IDH1-mutated glioblastomas, and IDH1 mutation status accounts for the unfavorable prognostic effect of higher age: implications for classification of gliomas. Acta Neuropathol (Berl) 120:707–718
Darlix A, Deverdun J, Menjot de Champfleur N et al (2017) IDH mutation and 1p19q codeletion distinguish two radiological patterns of diffuse low-grade gliomas. J Neurooncol 133:37–45
Metellus P, Coulibaly B, Colin C et al (2010) Absence of IDH mutation identifies a novel radiologic and molecular subtype of WHO grade II gliomas with dismal prognosis. Acta Neuropathol (Berl) 120:719–729
Patterson DM, Padhani AR, Collins DJ (2008) Technology insight: water diffusion MRI–a potential new biomarker of response to cancer therapy. Nat Clin Pract Oncol 5:220–233
Zhang L, Min Z, Tang M et al (2017) The utility of diffusion MRI with quantitative ADC measurements for differentiating high-grade from low-grade cerebral gliomas: Evidence from a meta-analysis. J Neurol Sci 373:9–15
Hempel J-M, Bisdas S, Schittenhelm J et al (2017) In vivo molecular profiling of human glioma using diffusion kurtosis imaging. J Neurooncol 131:93–101
Xiong J, Tan W, Wen J et al (2016) Combination of diffusion tensor imaging and conventional MRI correlates with isocitrate dehydrogenase 1/2 mutations but not 1p/19q genotyping in oligodendroglial tumours. Eur Radiol 26:1705–1715
Tan WL, Huang WY, Yin B et al (2014) Can diffusion tensor imaging noninvasively detect IDH1 gene mutations in astrogliomas? A retrospective study of 112 cases. AJNR Am J Neuroradiol 35:920–927
Han X, Suo S, Sun Y et al (2017) Apparent diffusion coefficient measurement in glioma: Influence of region-of-interest determination methods on apparent diffusion coefficient values, interobserver variability, time efficiency, and diagnostic ability. J Magn Reson Imaging JMRI 45:722–730
Pipe J (2009) Chapter 2 - pulse sequences for diffusion-weighted MRI. In: Johansen-Berg H, Behrens TEJ (eds) Diffusion MRI. Academic Press, San Diego, pp 11–35
Jenkinson M, Beckmann CF, Behrens TEJ et al (2012) FSL. NeuroImage 62:782–790
Yushkevich PA, Piven J, Hazlett HC et al (2006) User-guided 3D active contour segmentation of anatomical structures: significantly improved efficiency and reliability. NeuroImage 31:1116–1128
Jenkinson M, Bannister P, Brady M, Smith S (2002) Improved optimization for the robust and accurate linear registration and motion correction of brain images. NeuroImage 17:825–841
Jenkinson M, Smith S (2001) A global optimisation method for robust affine registration of brain images. Med Image Anal 5:143–156
Seiz M, Freyschlag CF, Schenkel S et al (2011) Management of patients with low-grade gliomas - a survey among German neurosurgical departments. Cent Eur Neurosurg 72:186–191
Khan OH, Mason W, Kongkham PN et al (2016) Neurosurgical management of adult diffuse low grade gliomas in Canada: a multi-center survey. J Neurooncol 126:137–149
Karavaeva E, Harris RJ, Leu K et al (2015) Relationship between [18F]FDOPA PET uptake, apparent diffusion coefficient (ADC), and proliferation rate in recurrent malignant gliomas. Mol Imaging Biol 17:434–442
Fudaba H, Shimomura T, Abe T et al (2014) Comparison of multiple parameters obtained on 3T pulsed arterial spin-labeling, diffusion tensor imaging, and MRS and the Ki-67 labeling index in evaluating glioma grading. AJNR Am J Neuroradiol 35:2091–2098
Ellingson BM, Malkin MG, Rand SD et al (2010) Validation of functional diffusion maps (fDMs) as a biomarker for human glioma cellularity. J Magn Reson Imaging JMRI 31:538–548
Higano S, Yun X, Kumabe T et al (2006) Malignant astrocytic tumors: clinical importance of apparent diffusion coefficient in prediction of grade and prognosis. Radiology 241:839–846
Miloushev VZ, Chow DS, Filippi CG (2015) Meta-analysis of diffusion metrics for the prediction of tumor grade in gliomas. AJNR Am J Neuroradiol 36:302–308
Zulfiqar M, Yousem DM, Lai H (2013) ADC values and prognosis of malignant astrocytomas: does lower ADC predict a worse prognosis independent of grade of tumor? A meta-analysis. AJR Am J Roentgenol 200:624–629
Cui Y, Ma L, Chen X et al (2014) Lower apparent diffusion coefficients indicate distinct prognosis in low-grade and high-grade glioma. J Neurooncol 119:377–385
Khayal IS, Vandenberg SR, Smith KJ et al (2011) MRI apparent diffusion coefficient reflects histopathologic subtype, axonal disruption, and tumor fraction in diffuse-type grade II gliomas. Neuro Oncol 13:1192–1201
Tozer DJ, Jäger HR, Danchaivijitr N et al (2007) Apparent diffusion coefficient histograms may predict low-grade glioma subtype. NMR Biomed 20:49–57
Clark K, Voronovich Z, Horbinski C (2013) How molecular testing can help (and hurt) in the workup of gliomas. Am J Clin Pathol 139:275–288
Leu K, Ott GA, Lai A et al (2017) Perfusion and diffusion MRI signatures in histologic and genetic subtypes of WHO grade II-III diffuse gliomas. J Neurooncol. https://doi.org/10.1007/s11060-0172506-9
Arponen O, Arponent O, Sudah M et al (2015) Diffusion-weighted imaging in 3.0 Tesla breast MRI: diagnostic performance and tumor characterization using small subregions vs. whole tumor regions of interest. PloS One 10:e0138702
Xing Z, Yang X, She D et al (2017) Noninvasive assessment of IDH mutational status in World Health Organization grade II and III astrocytomas using DWI and DSC-PWI combined with conventional MR imaging. AJNR Am J Neuroradiol 38:1138–1144
Lee S, Choi SH, Ryoo I et al (2015) Evaluation of the microenvironmental heterogeneity in high-grade gliomas with IDH1/2 gene mutation using histogram analysis of diffusion-weighted imaging and dynamic-susceptibility contrast perfusion imaging. J Neurooncol 121:141–150
Yamashita K, Hiwatashi A, Togao O et al (2016) MR imaging-based analysis of glioblastoma multiforme: estimation of IDH1 mutation status. AJNR Am J Neuroradiol 37:58–65
Kickingereder P, Sahm F, Radbruch A et al (2015) IDH mutation status is associated with a distinct hypoxia/angiogenesis transcriptome signature which is non-invasively predictable with rCBV imaging in human glioma. Sci Rep 5:16238
Andronesi OC, Rapalino O, Gerstner E et al (2013) Detection of oncogenic IDH1 mutations using magnetic resonance spectroscopy of 2-hydroxyglutarate. J Clin Invest 123:3659–3663
Lev MH, Ozsunar Y, Henson JW et al (2004) Glial tumor grading and outcome prediction using dynamic spin-echo MR susceptibility mapping compared with conventional contrast-enhanced MR: confounding effect of elevated rCBV of oligodendrogliomas [corrected]. AJNR Am J Neuroradiol 25:214–221
Funding
This study received no specific funding, but was supported by the National Institute for Health Research University College London Biomedical Research Centre.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Guarantor
The scientific guarantor of this publication is Stefanie Thust.
Conflict of interest
The authors of this manuscript declare no relationships with any companies whose products or services may be related to the subject matter of the article.
Statistics and biometry
One of the authors (C.T. MD PhD MSc Medical Statistics) has significant statistics expertise.
Informed consent
Written informed consent was waived by the institutional review board.
Ethical approval
Institutional review board approval was obtained.
Methodology
• retrospective
• diagnostic study/observational
• performed at one institution
Rights and permissions
Open Access This article is distributed under the terms of the Creative Commons Attribution 4.0 International License (http://creativecommons.org/licenses/by/4.0/), which permits unrestricted use, distribution, and reproduction in any medium, provided you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made.
About this article
Cite this article
Thust, S.C., Hassanein, S., Bisdas, S. et al. Apparent diffusion coefficient for molecular subtyping of non-gadolinium-enhancing WHO grade II/III glioma: volumetric segmentation versus two-dimensional region of interest analysis. Eur Radiol 28, 3779–3788 (2018). https://doi.org/10.1007/s00330-018-5351-0
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00330-018-5351-0