Abstract
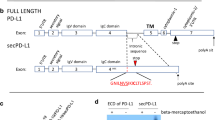
Therapeutic blockade of the PD-1/PD-L1 axis is recognized as an effective treatment for numerous cancer types. However, only a subset of patients respond to this treatment, warranting a greater understanding of the biological mechanisms driving immune evasion via PD-1/PD-L1 signaling and other T-cell suppressive pathways. We previously identified a head and neck squamous cell carcinoma with human papillomavirus integration in the PD-L1 locus upstream of the transmembrane domain-encoding region, suggesting expression of a truncated form of PD-L1 (Parfenov et al., Proc Natl Acad Sci USA 111(43):15544–15549, 2014). In this study, we extended this observation by performing a computational analysis of 33 other cancer types as well as human cancer cell lines, and identified additional PD-L1 isoforms with an exon 4 enrichment expressed in 20 cancers and human cancer cell lines. We demonstrate that cancer cell lines with high expression levels of exon 4-enriched PD-L1 generate a secreted form of PD-L1. Further biochemical studies of exon 4-enriched PD-L1 demonstrated that this form is secreted and maintains the capacity to bind PD-1 as well as to serve as a negative regulator on T cell function, as measured by inhibition of IL-2 and IFNg secretion. Overall, we have demonstrated that truncated forms of PD-L1 exist in numerous cancer types, and have validated that truncated PD-L1 can be secreted and negatively regulate T cell function.




Similar content being viewed by others
Abbreviations
- ATCC:
-
American type culture collection
- CCLE:
-
Cancer cell line encyclopedia
- CST:
-
Cell signaling technology
- EBV:
-
Epstein–Barr virus
- ELISA:
-
Enzyme linked immunosorbent assay
- FPKM:
-
Fragments per kilobase of transcript per million mapped reads
- GTeX:
-
Genotype-Tissue Expression
- HA:
-
Hemagglutinin
- HNSCC:
-
Head and neck squamous cell carcinoma
- HPV:
-
Human papillomavirus
- HRP:
-
Horseradish Peroxidase
- NCI:
-
National Cancer Institute
- PD-L1:
-
Programmed death-ligand-1
- PHA:
-
phytohemagglutinin
- PSG:
-
Penicillin/streptomycin/glutamine
- RIPA:
-
Radioimmunoprecipitation assay
- RT:
-
Room temperature
- TCA:
-
Trichloroacetic acid
- TCGA:
-
The Cancer Genome Atlas
- TMB:
-
3,3′,5,5′-Tetramethylbenzidine
References
Mahoney KM, Rennert PD, Freeman GJ (2015) Combination cancer immunotherapy and new immunomodulatory targets. Nat Rev Drug Discov 14(8):561–584
Postow MA, Callahan MK, Wolchok JD (2015) Immune checkpoint blockade in cancer therapy. J Clin Oncol 33(17):1974–1982
Chen Z et al (2014) Non-small-cell lung cancers: a heterogeneous set of diseases. Nat Rev Cancer 14(8):535–546
Pardoll DM (2012) The blockade of immune checkpoints in cancer immunotherapy. Nat Rev Cancer 12(4):252–264
Freeman GJ et al (2000) Engagement of the PD-1 immunoinhibitory receptor by a novel B7 family member leads to negative regulation of lymphocyte activation. J Exp Med 192(7):1027–1034
Keir ME et al (2008) PD-1 and its ligands in tolerance and immunity. Annu Rev Immunol 26:677–704
Gillison ML et al (2000) Evidence for a causal association between human papillomavirus and a subset of head and neck cancers. J Natl Cancer Inst 92(9):709–720
Yim EK, Park JS (2005) The role of HPV E6 and E7 oncoproteins in HPV-associated cervical carcinogenesis. Cancer Res Treat 37(6):319–324
Gulley ML (2015) Genomic assays for Epstein–Barr virus-positive gastric adenocarcinoma. Exp Mol Med 47:e134
Network CGAR (2014) Comprehensive molecular characterization of gastric adenocarcinoma. Nature 513(7517):202–209
Green MR et al (2012) Constitutive AP-1 activity and EBV infection induce PD-L1 in Hodgkin lymphomas and posttransplant lymphoproliferative disorders: implications for targeted therapy. Clin Cancer Res 18(6):1611–1618
Lyford-Pike S et al (2013) Evidence for a role of the PD-1:PD-L1 pathway in immune resistance of HPV-associated head and neck squamous cell carcinoma. Cancer Res 73(6):1733–1741
Parfenov M et al (2014) Characterization of HPV and host genome interactions in primary head and neck cancers. Proc Natl Acad Sci USA 111(43):15544–15549
Ojesina AI et al (2014) Landscape of genomic alterations in cervical carcinomas. Nature 506(7488):371–375
Kataoka K, Shiraishi Y, Takeda Y, Sakata S, Matsumoto M, Nagano S, Maeda T, Nagata Y (2016) Aberrant PD-L1 expression through 3′-UTR disruption in multiple cancers. Nature 537:402
Frigola X et al (2011) Identification of a soluble form of B7-H1 that retains immunosuppressive activity and is associated with aggressive renal cell carcinoma. Clin Cancer Res 17(7):1915–1923
Zheng Z et al (2014) Level of circulating PD-L1 expression in patients with advanced gastric cancer and its clinical implications. Chin J Cancer Res 26(1):104–111
Zhang J et al (2015) Circulating PD-L1 in NSCLC patients and the correlation between the level of PD-L1 expression and the clinical characteristics. Thorac Cancer 6(4):534–538
Grossman RL et al (2016) Toward a shared vision for cancer genomic data. N Engl J Med 375(12):1109–1112
Pertea M et al (2016) Transcript-level expression analysis of RNA-seq experiments with HISAT, StringTie and Ballgown. Nat Protoc 11(9):1650–1667
Pertea M et al (2015) StringTie enables improved reconstruction of a transcriptome from RNA-seq reads. Nat Biotechnol 33(3):290–295
Weinstein JN et al (2013) The cancer genome atlas pan-cancer analysis project. Nat Genet 45(10):1113–1120
Consortium G (2013) The genotype-tissue expression (GTEx) project. Nat Genet 45(6):580–585
Collado-Torres L et al (2017) Reproducible RNA-seq analysis using recount2. Nat Biotechnol 35(4):319–321
Zhou J et al (2017) Soluble PD-L1 as a biomarker in malignant melanoma treated with checkpoint blockade. Cancer Immunol Res 5(6):480–492
Keir ME, Francisco LM, Sharpe AH (2007) PD-1 and its ligands in T-cell immunity. Curr Opin Immunol 19(3):309–314
Butte MJ et al (2008) Interaction of human PD-L1 and B7-1. Mol Immunol 45(13):3567–3572
Haile ST et al (2013) Soluble CD80 restores T cell activation and overcomes tumor cell programmed death ligand 1-mediated immune suppression. J Immunol 191(5):2829–2836
Schneider CA, Rasband WS, Eliceiri KW (2012) NIH Image to ImageJ: 25 years of image analysis. Nat Methods 9(7):671–675
Cheng S et al (2015) PD-L1 gene polymorphism and high level of plasma soluble PD-L1 protein may be associated with non-small cell lung cancer. Int J Biol Markers 30(4):e364–e368
Hendrickx W et al (2017) Identification of genetic determinants of breast cancer immune phenotypes by integrative genome-scale analysis. Oncoimmunology 6(2):e1253654
Balar AV et al (2017) Atezolizumab as first-line treatment in cisplatin-ineligible patients with locally advanced and metastatic urothelial carcinoma: a single-arm, multicentre, phase 2 trial. Lancet 389(10064):67–76
Rosenberg JE et al (2016) Atezolizumab in patients with locally advanced and metastatic urothelial carcinoma who have progressed following treatment with platinum-based chemotherapy: a single-arm, multicentre, phase 2 trial. Lancet 387(10031):1909–1920
Barretina J et al (2012) The cancer cell line encyclopedia enables predictive modelling of anticancer drug sensitivity. Nature 483(7391):603–607
Chen Y et al (2011) Development of a sandwich ELISA for evaluating soluble PD-L1 (CD274) in human sera of different ages as well as supernatants of PD-L1 + cell lines. Cytokine 56(2):231–238
Frigola X et al (2012) Soluble B7-H1: differences in production between dendritic cells and T cells. Immunol Lett 142(1–2):78–82
Theodoraki MN et al (2018) Clinical significance of PD-L1. Clin Cancer Res 24(4):896–905
Acknowledgements
The authors would like to acknowledge the Dana-Farber Cancer Institute Flow Cytometry Core (Suzan Lazo-Kallanian, John Daley), and UT Southwestern Children’s Research Institute Flow Cytometry Core for help with flow cytometry. The authors would also like to thank Gordon Freeman for PD-L1 antibodies.
Funding
This work was supported by the NCI R01 CA205150, CA196932, and K08 CA163677 grants, as well as the Starr Consortium for Cancer Research and Stand up to Cancer awards to Peter Hammerman. This work was also supported by the Young Investigator Award from the International Association for the Study of Lung Cancer, Career Enhancement Award (5P50CA070907) through the National Institutes of Health, and Cancer Prevention and Research Institute of Texas Scholar Award (RR160080) to Esra Akbay. Venkat Malladi was supported by the Cancer Prevention and Research Institute of Texas award (RP150596).
Author information
Authors and Affiliations
Contributions
NBH and VSM contributed equally to experimental design, data collection and analysis, and manuscript writing. YH and EMB helped with data collection and analysis. SSF and CSP helped with bioinformatics. SK, SM, NS, K-KW and GD aided with experimental design and provided reagents. PSH and EAA helped with experimental design, data collection, providing reagents, and manuscript writing and editing.
Corresponding author
Ethics declarations
Conflict of interest
The authors have no relevant conflicts to disclose.
Informed consent
Informed consent was obtained from blood donors from the Brigham and Women’s Blood Bank under the approved IRB protocol 02-180.
Cell line authentication
HEK293T cells were purchased from ATCC, and RKO, CAL62, and RERF-LC-Ad1 cells were obtained from the CCLE [34] through the Broad Institute of Massachusetts Institute of Technology and Harvard. All cells were used within 6 months of initial passage and not further authenticated.
Additional information
Poster/abstract publication in Cancer Immunology Research, 2017; 5 (3 Suppl); Abstract nr A56; Proceedings of the American Association for Cancer Research Special Conference on Tumor Immunology and Immunotherapy; 2016 Oct 21; Boston, MA, USA.
This paper is published together with the following paper https://doi.org/10.1007/s00262-018-2282-1.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Hassounah, N.B., Malladi, V.S., Huang, Y. et al. Identification and characterization of an alternative cancer-derived PD-L1 splice variant. Cancer Immunol Immunother 68, 407–420 (2019). https://doi.org/10.1007/s00262-018-2284-z
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00262-018-2284-z