Abstract
The variant strains of porcine epidemic diarrhea virus (PEDV) severely threaten the pig industry worldwide and cause up to 100% mortality in suckling piglets. It is critically important and urgent to develop tools for detection of PEDV infection. In this study, we developed six monoclonal antibodies (mAbs) targeting N protein of PEDV and analyzed their applications on enzyme-linked immunosorbent assay (ELISA), indirect immunofluorescence assay (IFA), western blot assay, and flow cytometry assay. The results demonstrated that all these six mAbs were IgG1 isotype and κ chain. Among these six mAbs, 3F12 recognizes a linear epitope (VAAVKDALKSLGI) while the other five mAbs recognize different conformational epitopes formed by a specific peptide fragment or the full length of N protein. The functional analysis showed that all these six mAbs were applicable to ELISA, western blot, IFA, and flow cytometry assay. In conclusion, we developed six mAbs against PEDV-N protein to facilitate the early detection of PEDV infection using ELISA, western blot, IFA, and flow cytometry.
Similar content being viewed by others
Introduction
Porcine epidemic diarrhea virus (PEDV), a member of the Alphacoronavirus genus from the Coronaviradae family, mainly infects swine enterocytes and causes intestinal atrophy, dehydration, and diarrhea in pigs (Chasey and Cartwright 1978; Huang et al. 2013). Generally, PEDV is considered as a food-borne and water-borne pathogen and mainly transmitted through the fecal–oral route. However, recent studies showed that PEDV can also be transmitted through the air (Alvarez et al. 2016; Beam et al. 2015; Li et al. 2018). Even though PEDV has been found in pig herds for more than 40 years, it did not cause severe outbreaks and significant economic losses previously. However, new PEDV variant strains with extremely high virulence to piglets had reemerged in pig farms of China since late 2011 (Li et al. 2012; Sun et al. 2012) and had been found in the Americas, the European Union, and Asian countries since then. The reemerged PEDV strains caused up to 100% mortality in suckling piglets younger than 1 week due to the high virulence of these variants and the lack of a mature immune system in piglets (Stevenson et al. 2013).
Similar to other members in the Coronaviradae family, PEDV mainly contains four structural proteins: spike protein (S), envelope protein (E), membrane protein (M), and nucleocapsid protein (N) (Kocherhans et al. 2001). Among them, the N protein packages the PEDV genomic RNA into a helical ribonucleoprotein (RNP) and plays a fundamental role during virion assembly through its interactions with the viral genome and membrane protein M (Egberink et al. 1988). It also plays an important role in enhancing the efficiency of subgenomic viral RNA transcription as well as viral replication (Egberink et al. 1988). Additionally, N protein is highly conserved between different PEDV strains (Song and Park 2012). Therefore, the N protein can be used as a target for early detection of PEDV infection and evaluation of immunization efficacy raised by PEDV live vaccine.
To facilitate comprehensive research of PEDV infection and immune responses, we prepared a series of monoclonal antibodies (mAbs) targeting PEDV-N protein in this study. The functions of these mAbs were validated by enzyme-linked immunosorbent assay (ELISA), indirect immunofluorescence assay (IFA), western blot assay, and flow cytometry.
Materials and methods
Cells, virus, and mice
A mouse myeloma cell line (SP2/0, ATCC CRL-1581) was cultured in DMEM basic medium containing 20% fetal bovine serum (FBS) in a 37 °C CO2 incubator while African green monkey kidney cells (Vero-E6, ATCC CRL-1586) were maintained in DMEM medium containing 10% FBS. All culture media were supplied with 0.1 mg/ml streptomycin and 100 IU/ml penicillin. The PEDV LJX01/2014 strain was isolated from a pig farm and propagated in our laboratory into a cell-adapted virus. It is publicly available through the corresponding authors upon request. Six-week-old SPF BALB/c mice were housed in isolators located at Lanzhou Veterinary Research Institute (LVRI) animal facility. All animal works were done according to the animal care protocol which was approved by the LVRI experimental animal committee.
Expression of full-length and truncated PEDV-N proteins
The full-length open reading frame of N protein from PEDV LJX01/2014 strain (GenBank accession number MK252703) was amplified using a standard RT-PCR procedure with primers N-F: 5′-CGGGATCCATGGCTTCTGTCAG-3′ (contains a BamH I digestible site) and N-R: 5′-CGAAGCTTATTTCCTGTGTCGAAGAT-3′ (attached a Hind III site), and subcloned into the EcoR I and BamH I sites of the pET-30(a) vector (Novagen). The recombinant plasmid was verified by PCR amplification with the universal primer set, digestion with restriction enzymes, and DNA sequencing. The positive recombinant plasmid was named as pET-N. The pET-N was then transformed into competent cells BL21(DE3) for expression of a full-length protein under the induction with 0.5 mM IPTG. The expressed proteins were identified with 10% SDS-PAGE and confirmed by western blot assay using His-tag mAb as a primary antibody and HRP-conjugated goat anti-mouse IgG as a secondary antibody. The recombinant N protein was purified by nickel column affinity chromatography.
To map the epitopes of yield PEDV mAbs, the gene of PEDV-N protein was truncated into five fragments with 60-bp overlap between two fragments and cloned into pET-30(a) vector. The detailed primers amplifying these fragments are listed in Supplementary Table S1. The truncated N protein fragments were expressed in BL21(DE3) Escherichia coli cells as mentioned above.
Production of PEDV-N mAbs
Four 6-week-old female BALB/c mice, provided by the Experimental Animal Center of Harbin Veterinary Research Institute, CAAS, were subcutaneously immunized with 100 μg purified N protein mixed with 30% ISA adjuvant. Two weeks post-primary immunization, the mice were boosted twice with the same dose of N protein at a 2-week interval. Serum samples were collected for monitoring the antibody responses by indirect ELISA coated with recombinant N protein. Three days after the final vaccination, the mice were euthanized to harvest the spleen. The splenocyte single-cell suspension was prepared and fused with SP2/0 cell in 96-well plates at a ratio between 5:1 and 10:1 using polyethylene glycol (PEG 4000, Sigma). Hybridoma cells were cultured in HAT DMEM medium supplied with 20% FBS and antibiotics. Indirect ELISA coating with purified N protein was employed to screen the antibody-secreting hybridomas. The positive hybridoma cells were subcloned into a new 96-well plate by limiting dilutions. After three-round subcloning, the hybridomas stably secreting antibody were determined. For the preparation of ascites of mAb, a total of 106 hybridoma cells of each mAb were injected into the mouse peritoneal cavity. The ascites was collected with a sterile syringe daily 1 week after injection of hybridoma cells.
Enzyme-linked immunosorbent assay
Each well of an ELISA plate was coated with 200 ng purified N protein in coating buffer overnight at 4 °C then blocked with 5% skim milk for 2 h at 37 °C. The cell culture supernatant of hybridomas was added to each well as a primary antibody and incubated at 37 °C for 1 h. After three washes, 1:2000 diluted HRP-conjugated goat anti-mouse IgG (H+L) was added to wells and incubated at 37 °C for 1 h. The 3,3′,5,5′-tetramethylbenzidine (TMB) was subsequently added to all wells and incubated in the dark for approximately 10 min and stopped with 2 N H2SO4. The absorbance values were scanned at OD450nm using a microplate reader.
To identify the subtypes of mAbs obtained from this research, SBA Clonotyping System-HRP (Southern Biotech) was employed. Briefly, purified PEDV-N protein was used as coating antigen and prepared mAbs were used as primary antibodies for a conventional indirect ELISA assay. The HRP-conjugated goat anti-mouse Ig, IgG1, IgG2a, IgG2b, IgM, IgG3, IgA, kappa, and lambda were used as secondary antibodies to bind with primary antibody respectively.
For epitope mapping, the truncated PEDV-N protein fragments or synthesized peptides were used as coating antigen for indirect ELISA assay. All other procedures were the same as mentioned above.
Indirect immunofluorescence assay
Vero-E6 cells adhering on the glass slide in a 24-well plate were inoculated with PEDV LJX01/2014 strain at MOI = 0.01 and cultured for 24 h in a CO2 incubator at 37 °C. Then, the cells were fixed with 4% paraformaldehyde for 20 min and treated with 0.1% Triton X-100 for 15 min at room temperature. Subsequently, the cells were blocked with 5% BSA for 1 h at 37 °C. After three washes with PBS, diluted mAbs produced in this study were used as a primary antibody for overnight incubation at 4 °C. Then, 1:5000 diluted AF488-labeled goat anti-mouse IgG was added for 1-h incubation in the dark at 37 °C. The nucleus was stained with DAPI for 3–5 min. Finally, the stained slides were observed under a Vert-A1 fluorescence microscope (Zeiss).
Western blotting
The protein samples were subjected to 10% SDS-PAGE electrophoresis and then transferred onto the 0.45-μm PVDF membrane. Following blocking with 5% skim milk for 2 h at room temperature, the membrane was incubated with primary antibody overnight at 4 °C. After three washes with PBST, HRP-conjugated goat anti-mouse IgG was added to the membrane for 1-h incubation at 37 °C. The positive bands were visualized by electrochemiluminescence (ECL) reagent and developed on films.
For verifying the expression of recombinant proteins, the expressed or purified PEDV-N proteins (full length or truncated) were used as detecting proteins and His-tag mAb was served as the primary antibody. To identify the application of prepared mAbs on western blot analysis, cell lysates originating from PEDV-infected Vero-E6 cells were used as detecting proteins while the prepared mAbs were used as primary antibodies. Additionally, the β-actin was used as an internal control.
Flow cytometry analysis
The single-cell suspension was prepared from PEDV-infected Vero-E6 cells and stained with either prepared mAbs or mouse IgG1 isotype control for 30 min on ice. After three washes, the cells were incubated with AF647-labeled goat anti-mouse IgG. All samples were resuspended in PBS and a BD Accuri C6 flow cytometer was used for detection. The flow cytometry data were analyzed using the FlowJo software.
Statistical analysis
Data were analyzed and processed by GraphPad Prism 6.0 software. Student’s t test with two-tailed distribution and two-sample unequal variance was performed to determine statistical significance in pairwise comparisons.
Results
Expression and purification of recombinant PEDV-N protein
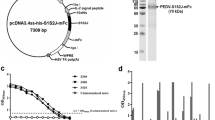
To express PEDV-N protein, the ORF of the N gene was firstly amplified with RT-PCR and cloned into pET-30(a) vector (Fig. 1a). The recombinant plasmid was verified by enzyme digestion with BamH I and Kpn I, and PCR amplification with T7 and T-term universal primer set. The results showed the right sizes for both digestion and amplification verification (Fig. 1a). The recombinant plasmid was also subjected to DNA sequencing analysis and the results demonstrated that the N gene was successfully cloned into pET-30(a) vector and no mutant was found (data not shown).
Expression and purification of recombinant PEDV-N protein. a The ORF of PEDV-N protein was amplified by RT-PCR and constructed into pET-30(a) vector. The plasmid of recombinant pET-N was extracted and identified by restriction endonuclease digestion and PCR amplification. b The recombinant plasmid pET-N was transformed into E. coli BL21(DE3) for protein expression. Recombinant PEDV-N protein was purified through Ni2+ column affinity chromatography. All protein samples were analyzed by SDS-PAGE and visualized by Coomassie Brilliant Blue staining. c The expressed and purified recombinant PEDV-N protein was identified by western blot using anti-His-tag primary antibody
The recombinant plasmid pET-N was then transformed into BL21(DE3) E. coli to express the target protein. SDS-PAGE results demonstrated that a specific protein band at approximately 70 kD was observed, compared with the expression products from pET-30(a) vector (Fig. 1b). The recombinant PEDV-N protein was purified with nickel column affinity chromatography and showed the recombinant PEDV-N in high purity was obtained (Fig. 1b). Western blot analysis using 6× His-tag mAb demonstrated the presence of recombinant PEDV-N protein (Fig. 1c). All these results indicated that the recombinant PEDV-N protein was successfully expressed and can be used to immunize mice for preparation of PEDV mAbs.
Preparation and characterization of PEDV-N mAbs
The BALB/c mice were immunized three times with recombinant PEDV-N protein with 2-week intervals. When the serum antibody level against PEDV-N protein reached the platform phase, the mice were sacrificed and splenocytes were isolated and fused with SP2/0 cells. Cell culture medium was measured by ELISA to screen the antibody-secreting hybridoma cells. The cells in positive wells were subjected to three-round subcloning with limited dilution strategy. Eventually, six mAb-secreting hybridoma strains were obtained and named 2D3, 3F12, 4A3, 5F3, 5F6, and 7G3 respectively. These six hybridoma cell strains were subsequently injected into mouse peritoneal cavity to produce mAbs in the ascites. The isotype of prepared mAbs was then analyzed by SBA Clonotyping System. The results demonstrated that the heavy chains of all six prepared mAbs against PEDV-N were IgG1 (Fig. 2a) while all the light chains were κ a chain (Fig. 2b). The antibody titer of the collected ascites was titrated by indirect ELISA with a series of dilutions. The results showed that most mAbs prepared in this study can be used in 1:16,000 dilution and the 2D3 can be diluted at least 4000-fold (Fig. 2b). The purified mAb 5F3 was selected to measure its affinity using the method reported by Friguet et al. (1985), and it turned out that the5F3 had a high affinity at 6.69 × 10−10 M (Supplementary Table S2).
Preparation and identification of PEDV-N mAbs. BALB/c mice were immunized by purified PEDV-N protein three times with 2-week intervals. Splenocytes prepared from immunized mice were fused with SP2/0 cell. The hybridoma cells were subjected to three-cycle subcloning. A total of six hybridoma cell lines secreting PEDV-N antibodies were obtained. The a heavy chain and b light chain of these 6 prepared mAbs were identified by ELISA using an SBA Clonotyping System. c Mouse ascites producing PEDV-N mAbs were prepared and subjected to a series of dilutions for antibody titration by ELISA
Epitope mapping of prepared mAbs
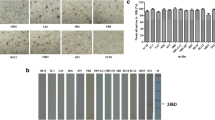
To determine the fine epitopes recognized by these prepared mAbs, the full-length ORF of PEDV-N gene was truncated into five segments with 60-bp overlap between neighboring parts (Fig. 3a) and constructed into pET-30(a) vector for expression of five truncated proteins. The result showed that all five truncated N proteins were successfully expressed in BL21(DE3) E. coli (Supplementary Fig. S1). The truncated PEDV-N proteins were used as coating antigen in an indirect ELISA assay to identify the locations of epitopes recognized by these mAbs. The results demonstrated that 3F12 and 7G3 had strong binding activity to PEDV N181-298 fragment while the 2D3 recognizes N271-379 (Fig. 3a). However, the remaining three mAbs, 4A3, 5F3, and 5F6, did not show any reactivity to all fragments except for the full-length PEDV-N protein (Fig. 3a), indicating these three mAbs may recognize conformational epitopes.
Epitope mapping of prepared mAbs. a The full-length PEDV-N protein was truncated into 5 segments with 20-amino acid overlap between neighboring parts. These 5 truncated fragments were expressed in E. coli expression system and used as coating antigens of ELISA to determine the epitope-located region recognized by 2D3, 3F12, 4A3, 5F3, 5F6, and 7G3 mAbs. b The fragment of PEDV N181-379 was divided into 9 peptides with 20-amino acid overlap. These 9 peptides were synthesized and used as coating antigen of ELISA to further map the epitope locations of 2D3, 3F12, and 7G3. c A panel of shortened peptides covering the whole or part of PEDV N221-240 fragment were synthesized and used as coating antigens of ELISA to determine the fine epitope of 7G3 and 3F12
Next, we synthesized a panel of 40-amino acid peptides covering the PEDV N181-379 fragment with 20-amino acid overlap between neighboring peptides to further map the epitopes recognized by 2D3, 3F12, and 7G3 (Fig. 3b). The synthesized peptides were used as coating antigens to measure the reactivity with these mAbs by indirect ELISA. The results demonstrated that 3F12 and 7G3 showed strong reactivity with both N201-240 and N221-260 peptides (Fig. 3b), indicating their epitopes may be located in the N221-240 fragment. However, 2D3 did not react with any of these synthesized peptides (Fig. 3b), implying that 2D3 should recognize a conformational epitope formed by N271-379 fragment.
In order to map the fine epitopes recognized by 3F12 and 7G3, another panel of peptides covering the N221-240 fragment was synthesized as indicated in Fig. 3c. Again, these peptides were used as coating antigens for indirect ELISA. The results demonstrated that the epitope recognized by 3F12 was N223-235, VAAKDALKSLGI (Fig. 3c). Unfortunately, 7G3 did not show any reactivity with any peptide in this panel, suggesting that 7G3 may recognize a conformational epitope formed by N221-240 fragment.
Applications of prepared mAbs
We next tested the applications of these prepared mAbs on ELISA. The purified PEDV-N protein was coated on an ELISA plate and all prepared mAbs were added to each coated well respectively as primary antibodies. After incubation with secondary antibody and developing, the readout of the ELISA plate demonstrated that all six mAbs can be used as primary antibodies to detect PEDV-N protein using the ELISA assay, with the best reactivity obtained from 2D3, 3F12, 5F6, and 7G3 strains (Fig. 4a). Additionally, we evaluate the sensitivity of detection of our prepared mAbs. The results showed that all six prepared mAbs could detect as little as 4.87–9.7 ng of purified PEDV-N protein (Supplementary Fig. S2).
Functional analysis of prepared mAbs. a The 6 prepared mAbs were used as the primary antibody in an indirect ELISA to test their reactivity with PEDV-N protein. Vero-E6 cells were inoculated with PEDV LJX01/2014 strain at MOI = 0.01 and harvested at 24 hpi. b The cell lysate was employed to determine the reactivity with these 6 prepared mAbs in western blot assay. c PEDV-infected or mock-infected Vero-E6 cells were fixed with 4% paraformaldehyde, treated with 0.1% Triton X-100, and then incubated with 6 prepared mAbs respectively. After incubating with AF488-labeled secondary antibody, the cells were stained with DAPI and visualized under fluorescent microscopy. The figures were merged from AF488 staining and DAPI staining. d The single-cell suspension was prepared, stained with the 6 mAbs, respectively, and then AF647-labeled goat anti-mouse secondary antibody. Mouse IgG1 was used as an isotype control. The cells were resuspended in PBS and analyzed by a BD Accuri C6 flow cytometer
To verify whether these mAbs were applicable to western blot assay, IFA, and flow cytometry assay, Vero-E6 cells were inoculated with PEDV and harvested at 24 hpi. For western blot analysis, PEDV-infected or mock-infected cells were harvested and lysed with lysis buffer. Total proteins in cell lysates were firstly isolated in an SDS-PAGE gel and then blotted onto a PVDF film. The film was split by lanes and incubated in diluted mAbs respectively. The western blot results showed that all mAbs can be used to detect N protein in PEDV-infected Vero-E6 cells while no non-specific bands were found in the mock-infected cells (Fig. 4b). For IFA detection, the PEDV-infected cells were attached to a glass slide and stained with these prepared mAbs. The results revealed that all the mAbs worked very well in the IFA assay, without non-specific staining on mock-infected cells (Fig. 4c). For flow cytometry analysis, a single-cell suspension of PEDV-infected Vero-E6 cells was prepared and incubated in six diluted mAbs respectively while the mouse IgG1 was used as isotype control. The flow cytometry results manifested that all the prepared mAbs can be used in flow cytometry to detect the PEDV-infected cells (Fig. 4d). The purified 5F3 mAb was selected and titrated on PEDV-infected Vero-E6 cells by flow cytometry. The results demonstrated this mAb is workable for as little as 0.031 μg per reaction (Supplementary Fig. S3). Further flow cytometry analyses indicated that all these mAbs could also be used to detect the infection caused by a classical PEDV strain, CV777 (Supplementary Fig. S4). All these results implied that these six mAbs prepared in this study were functional and useful for analyzing PEDV infection.
Discussion
Early detection of PEDV infection is of great importance due to its severe impact on the swine industry worldwide. However, the detection methods targeting the spike protein of PEDV lack accuracy because mutations always occur in this protein. Currently, cumulative mutations of virulent PEDV strains have led to the change of their antigenicity, resulting in a decrease or failure in the efficacy of the vaccines which originated from the classical strain CV777 (Song et al. 2015). Additionally, PEDV is often coinfected with some other diarrheal viruses (Zhang et al. 2013; Zhao et al. 2016). Meanwhile, the clinical symptoms between the PEDV and other porcine diarrheal viruses share a high similarity (Lin et al. 2015; Wang et al. 2016). Therefore, it is critically important and urgent to develop tools for detection of PEDV infection.
The N protein is an ideal target for early detection of PEDV or other coronavirus infections due to the following features and functions (Li et al. 2005; Totura and Baric 2012). Firstly, the N protein is a phosphorylated protein that binds stably to the viral RNA to form a nucleocapsid of the helical structure, which is involved in the transcription and translation of the viral genome, thereby affecting the replication of the virus (Hiscox et al. 2001; Saif 1993; Yang et al. 2013; You et al. 2005). In addition, the N protein interacts with the M protein to form budding vesicles, which is involved in the release process of the virus. The PEDV-N protein was reported to inhibit the production of type I interferon and activate the NF-κB pathway to escape the host’s antiviral immunity (Ding et al. 2014). What’s more, the N protein can be transported between nucleoli and cells via nuclear export signals (NES) using the CRM1-dependent pathway (Shi et al. 2014). More importantly, the N protein is highly conserved between PEDV strains (Li et al. 2013; Song et al. 2015). We selected some nucleoproteins of representative viral strains from different swine coronaviruses to compare their homologies with our PEDV strain LJX01/2014. The results showed that all the homologies of nucleoprotein between selected coronaviruses and PEDV LJX01/2014 are less than 44% (Supplementary Table S3). In addition, we analyzed the nucleoprotein homologies between PEDV LJX.01/2014 and other representative PEDV strains (classical or virulent) isolated from different countries and years. The results demonstrated that their nucleoprotein homologies between selected PEDV strains and LJX01/2014 are above 97% (Supplementary Table S4). These results indicated that nucleoprotein is highly conserved within PEDV strains while varying between different genuses of coronavirus. As a result, the PEDV-N protein can be used as a candidate for early detection of PEDV infection in vivo and in vitro.
To develop specific tools to facilitate PEDV detection, we prepared PEDV-N protein in high purity and generated six mAbs targeting this protein. Among these six mAbs, 3F12 recognizes a linear epitope (VAAVKDALKSLGI) while the other five mAbs recognize different conformational epitopes formed by a specific peptide fragment or the full length of N protein. Xie et al. recently reported two N-terminal epitopes located within residues 58–85 had cross-reactivity between PEDV and TGEV (Xie et al. 2019). However, all six mAbs prepared in this study recognized the epitopes located between residues 201–379 of PEDV nucleoprotein. Therefore, the mAbs we generated might be specific to PEDV infection. The functional analysis indicated that all six mAbs were applicable to ELISA, western blot, IFA, and flow cytometry assay. In addition, the PEDV-neutralizing activity of these six mAbs was determined by antibody neutralization assay. Unfortunately, none of these six mAbs can neutralize PEDV, which was different from the neutralization effect obtained from the antibodies originating from PEDV-S protein (Li et al. 2017; Liu et al. 2017; Okda et al. 2017).
In conclusion, we developed six mAbs against PEDV-N protein to facilitate the early detection of PEDV infection using ELISA, western blot, IFA, and flow cytometry assay.
References
Alvarez J, Goede D, Morrison R, Perez A (2016) Spatial and temporal epidemiology of porcine epidemic diarrhea (PED) in the Midwest and Southeast regions of the United States. Prev Vet Med 123:155–160. https://doi.org/10.1016/j.prevetmed.2015.11.003
Beam A, Goede D, Fox A, McCool MJ, Wall G, Haley C, Morrison R (2015) A porcine epidemic diarrhea virus outbreak in one geographic region of the United States: descriptive epidemiology and investigation of the possibility of airborne virus spread. PLoS One 10(12):e0144818. https://doi.org/10.1371/journal.pone.0144818
Chasey D, Cartwright SF (1978) Virus-like particles associated with porcine epidemic diarrhoea. Res Vet Sci 25(2):255–256. https://doi.org/10.1016/S0034-5288(18)32994-1
Ding Z, Fang L, Jing H, Zeng S, Wang D, Liu L, Zhang H, Luo R, Chen H, Xiao S (2014) Porcine epidemic diarrhea virus nucleocapsid protein antagonizes beta interferon production by sequestering the interaction between IRF3 and TBK1. J Virol 88(16):8936–8945. https://doi.org/10.1128/jvi.00700-14
Egberink HF, Ederveen J, Callebaut P, Horzinek MC (1988) Characterization of the structural proteins of porcine epizootic diarrhea virus, strain CV777. Am J Vet Res 49(8):1320–1324
Friguet B, Chaffotte AF, Djavadi-Ohaniance L, Goldberg ME (1985) Measurements of the true affinity constant in solution of antigen-antibody complexes by enzyme-linked immunosorbent assay. J Immunol Methods 77(2):305–319. https://doi.org/10.1016/0022-1759(85)90044-4
Hiscox JA, Wurm T, Wilson L, Britton P, Cavanagh D, Brooks G (2001) The coronavirus infectious bronchitis virus nucleoprotein localizes to the nucleolus. J Virol 75(1):506–512. https://doi.org/10.1128/jvi.75.1.506-512.2001
Huang YW, Dickerman AW, Pineyro P, Li L, Fang L, Kiehne R, Opriessnig T, Meng XJ (2013) Origin, evolution, and genotyping of emergent porcine epidemic diarrhea virus strains in the United States. MBio 4(5):e00737–e00713. https://doi.org/10.1128/mBio.00737-13
Kocherhans R, Bridgen A, Ackermann M, Tobler K (2001) Completion of the porcine epidemic diarrhoea coronavirus (PEDV) genome sequence. Virus Genes 23(2):137–144. https://doi.org/10.1023/a:1011831902219
Li C, Li W, Lucio de Esesarte E, Guo H, van den Elzen P, Aarts E, van den Born E, Rottier PJM, Bosch BJ (2017) Cell attachment domains of the porcine epidemic diarrhea virus spike protein are key targets of neutralizing antibodies. J Virol 91(12). https://doi.org/10.1128/JVI.00273-17
Li W, Li H, Liu Y, Pan Y, Deng F, Song Y, Tang X, He Q (2012) New variants of porcine epidemic diarrhea virus, China, 2011. Emerg Infect Dis 18(8):1350–1353. https://doi.org/10.3201/eid1808.120002
Li Y, Wu Q, Huang L, Yuan C, Wang J, Yang Q (2018) An alternative pathway of enteric PEDV dissemination from nasal cavity to intestinal mucosa in swine. 9(1):3811 doi:https://doi.org/10.1038/s41467-018-06056-w
Li YH, Li J, Liu XE, Wang L, Li T, Zhou YH, Zhuang H (2005) Detection of the nucleocapsid protein of severe acute respiratory syndrome coronavirus in serum: comparison with results of other viral markers. J Virol Methods 130(1–2):45–50. https://doi.org/10.1016/j.jviromet.2005.06.001
Li Z, Chen F, Yuan Y, Zeng X, Wei Z, Zhu L, Sun B, Xie Q, Cao Y, Xue C, Ma J, Bee Y (2013) Sequence and phylogenetic analysis of nucleocapsid genes of porcine epidemic diarrhea virus (PEDV) strains in China. Arch Virol 158(6):1267–1273. https://doi.org/10.1007/s00705-012-1592-4
Lin CM, Gao X, Oka T, Vlasova AN, Esseili MA, Wang Q, Saif LJ (2015) Antigenic relationships among porcine epidemic diarrhea virus and transmissible gastroenteritis virus strains. J Virol 89(6):3332–3342. https://doi.org/10.1128/jvi.03196-14
Liu J, Shi H, Chen J, Zhang X, Ji Z, Yuan J, Shi D, Cao L, Zhu X, Dong H, Wang X, Zhang J, Feng L (2017) Neutralization of genotype 2 porcine epidemic diarrhea virus strains by a novel monoclonal antibody. Virology 507:257–262. https://doi.org/10.1016/j.virol.2017.04.026
Okda FA, Lawson S, Singrey A, Nelson J, Hain KS, Joshi LR, Christopher-Hennings J, Nelson EA, Diel DG (2017) The S2 glycoprotein subunit of porcine epidemic diarrhea virus contains immunodominant neutralizing epitopes. Virology 509:185–194. https://doi.org/10.1016/j.virol.2017.06.013
Saif LJ (1993) Coronavirus immunogens. Vet Microbiol 37(3–4):285–297. https://doi.org/10.1016/0378-1135(93)90030-B
Shi D, Lv M, Chen J, Shi H, Zhang S, Zhang X, Feng L (2014) Molecular characterizations of subcellular localization signals in the nucleocapsid protein of porcine epidemic diarrhea virus. Viruses 6(3):1253–1273. https://doi.org/10.3390/v6031253
Song D, Huang D, Peng Q, Huang T, Chen Y, Zhang T, Nie X, He H, Wang P, Liu Q, Tang Y (2015) Molecular characterization and phylogenetic analysis of porcine epidemic diarrhea viruses associated with outbreaks of severe diarrhea in piglets in Jiangxi, China 2013. PLoS One 10(3):e0120310. https://doi.org/10.1371/journal.pone.0120310
Song D, Park B (2012) Porcine epidemic diarrhoea virus: a comprehensive review of molecular epidemiology, diagnosis, and vaccines. Virus Genes 44(2):167–175. https://doi.org/10.1007/s11262-012-0713-1
Stevenson GW, Hoang H, Schwartz KJ, Burrough ER, Sun D, Madson D, Cooper VL, Pillatzki A, Gauger P, Schmitt BJ, Koster LG, Killian ML, Yoon KJ (2013) Emergence of porcine epidemic diarrhea virus in the United States: clinical signs, lesions, and viral genomic sequences. J Vet Diagn Investig 25(5):649–654. https://doi.org/10.1177/1040638713501675
Sun RQ, Cai RJ, Chen YQ, Liang PS, Chen DK, Song CX (2012) Outbreak of porcine epidemic diarrhea in suckling piglets, China. Emerg Infect Dis 18(1):161–163. https://doi.org/10.3201/eid1801.111259
Totura AL, Baric RS (2012) SARS coronavirus pathogenesis: host innate immune responses and viral antagonism of interferon. Curr Opin Virol 2(3):264–275. https://doi.org/10.1016/j.coviro.2012.04.004
Wang D, Fang L, Xiao S (2016) Porcine epidemic diarrhea in China. Virus Res 226:7–13. https://doi.org/10.1016/j.virusres.2016.05.026
Xie W, Ao C, Yang Y, Liu Y, Liang R, Zeng Z, Ye G, Xiao S, Fu ZF, Dong W, Peng G (2019) Two critical N-terminal epitopes of the nucleocapsid protein contribute to the cross-reactivity between porcine epidemic diarrhea virus and porcine transmissible gastroenteritis virus. J Gen Virol 100(2):206–216. https://doi.org/10.1099/jgv.0.001216
Yang Y, Zhang L, Geng H, Deng Y, Huang B, Guo Y, Zhao Z, Tan W (2013) The structural and accessory proteins M, ORF 4a, ORF 4b, and ORF 5 of Middle East respiratory syndrome coronavirus (MERS-CoV) are potent interferon antagonists. Protein Cell 4(12):951–961. https://doi.org/10.1007/s13238-013-3096-8
You J, Dove BK, Enjuanes L, DeDiego ML, Alvarez E, Howell G, Heinen P, Zambon M, Hiscox JA (2005) Subcellular localization of the severe acute respiratory syndrome coronavirus nucleocapsid protein. J Gen Virol 86(Pt 12):3303–3310. https://doi.org/10.1099/vir.0.81076-0
Zhang Q, Hu R, Tang X, Wu C, He Q, Zhao Z, Chen H, Wu B (2013) Occurrence and investigation of enteric viral infections in pigs with diarrhea in China. Arch Virol 158(8):1631–1636. https://doi.org/10.1007/s00705-013-1659-x
Zhao ZP, Yang Z, Lin WD, Wang WY, Yang J, Jin WJ, Qin AJ (2016) The rate of co-infection for piglet diarrhea viruses in China and the genetic characterization of porcine epidemic diarrhea virus and porcine kobuvirus. Acta Virol 60(1):55–61. https://doi.org/10.4149/av_2016_01_55
Funding
This work was supported by the National Key R&D Program of China (2016YFD0500103), the National Natural Science Foundation of China (31572498, 31702209), and the Elite Youth Program of CAAS.
Author information
Authors and Affiliations
Corresponding authors
Ethics declarations
Conflict of interest
The authors declare that they have no competing interests.
Ethical approval
All the mouse experimental procedures were approved by the Lanzhou Veterinary Research Institute experimental animal committee. All applicable international, national, and institutional guidelines for the care and use of animals were strictly followed.
Additional information
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Electronic supplementary material
ESM 1
(PDF 872 kb)
Rights and permissions
About this article
Cite this article
Yang, W., Chen, W., Huang, J. et al. Generation, identification, and functional analysis of monoclonal antibodies against porcine epidemic diarrhea virus nucleocapsid. Appl Microbiol Biotechnol 103, 3705–3714 (2019). https://doi.org/10.1007/s00253-019-09702-5
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00253-019-09702-5