Abstract
This review focusses on the energetics of protein translocation via the Sec translocation machinery. First we complement structural data about SecYEG’s conformational rearrangements by insight obtained from functional assays. These include measurements of SecYEG permeability that allow assessment of channel gating by ligand binding and membrane voltage. Second we will discuss the power stroke and Brownian ratcheting models of substrate translocation and the role that the two models assign to the putative driving forces: (i) ATP (SecA) and GTP (ribosome) hydrolysis, (ii) interaction with accessory proteins, (iii) membrane partitioning and folding, (iv) proton motive force (PMF), and (v) entropic contributions. Our analysis underlines how important energized membranes are for unravelling the translocation mechanism in future experiments.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Structural Insights into SecY Conformational States During Translocation
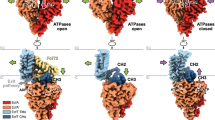
The Sec machinery is responsible for the reconstitution and translocation of many bacterial cytoplasmic, outer membrane and secretory proteins. The core element of the Sec translocation machinery is the heterotrimeric translocon SecYEG which resides in the cytoplasmic membrane (van den Berg et al. 2004). SecYEG has a striking homology with archaeal and eukaryotic analogues SecYEβ and Sec61αβγ (Bondar et al. 2010). All Sec translocons have a central pore that is closed for the passage of molecules in its resting state, due to the ring of hydrophobic amino acids (marked green in Fig. 1a) and the re-entrant loop TM2a also called the plug domain (marked yellow) (Saparov et al. 2007). The main translocation unit of the translocon SecY can be seen as a clamshell (blue and red halves) with a hinge region clamped by SecE, and the so-called lateral gate—composed of transmembrane helixes TM2b and TM7 on opposite sides (van den Berg et al. 2004). It was suggested that conformational transitions between the open and closed states can be seen as a rigid body movement of one-half against the other (Park et al. 2013).
a Crystal structure of the Methanococcus jannaschii SecY channel in its resting state (van den Berg et al. 2004) (PDB 1RH5). Left panel: channel viewed from the cytosol. Right panel: channel side view from the plasma membrane. b Closer view on the hydrophobic seal formed by the pore ring residues (green spheres). The pore ring of H. sapiens Sec61α is formed by 3 Ile, 2 Lue, 1 Val (right panel); and of M. jannaschii SecY by 4 Ile, 1 Leu, 1 Val (left panel). Complex of Sec61 with a translocation intermediate (right panel: engaged structure in red, signal sequence in cyan, PDB 3JC2) (Voorhees und Hegde 2016) as compared to the resting state of SecY (left panel: quiescent structure in grey, PDB 1RH5). (Color figure online)
Bacterial proteins can either be translocated via SecYEG, when still being translated by the ribosome (co-translationally) or after having been fully synthesized (post-translationally). Transmembrane proteins usually take the co-translational pathway where the protein is inserted into the lipid bilayer by the complex of the translating ribosome and the translocon. The secretory or outer membrane proteins that still contain their signal sequences (pre-proteins) use the post-translational pathway which requires a complex of SecY with the motor protein SecA (Junne et al. 2007; Denks et al. 2014). SecA was reported to interact with the substrate as it emerges from the ribosome (Huber et al. 2011). Co- and post-translational processes might even be intertwined, since SecA and the ribosome have overlapping binding sites on SecY and hence compete for binding (Kuhn et al. 2011).
To gain insight into how the translocon accommodates its substrates during translocation, we collected key structural information (Table 1). The structures can be grouped into those that show the resting (SecY alone), primed (with translocation partner bound such as SecA or empty ribosome), and engaged (in complex with translocation substrate) states of SecY. Since SecY engaged in translocation has only been captured in low resolution (except for the structure 5EUL (Li et al. 2016) mentioned below), Table 1 contains a mix of high- and low-resolution structures. We also included the RNC-Sec61 complex for comparison [pdb 3J46 (Voorhees et al. 2014)]. This structure is especially valuable because it captures the translocation intermediate without artificial cross-linking of the substrate to the translocon. SecY’s constriction zones regulate protein translocation. They consist of the hydrophobic ring for secretory proteins, and the lateral gate for transmembrane proteins. In addition to the width of these two important regulatory elements, the position of the plug domain—the third regulatory element—is of interest, because it governs the translocon’s permeability (Saparov et al. 2007). Table 1 summarizes current knowledge about all three regulatory elements:
-
(i)
In the RNC-Sec61, the plug has not been resolved, conceivably because it is mobile during translocation. To highlight plug mobility, Table 1 reports plug distances from two different reference points (see Table 1).
-
(ii)
The hydrophobic ring adopts the form of an ellipse with one axis being about twice as large as the other. It transforms into a more circular shape when the translocon is primed or engaged.
-
(iii)
Lateral gate opening tends to be minimal in the resting state. Different organisms such as M. jannaschii and T. thermophilus are almost identical in terms of lateral gate widths and “ring” shapes and sizes.
-
(iv)
The engaged structures either have a resolution which does not allow us to reliably distinguish between the conformational transitions in the constriction zone, or actually reproduce the resting state with astonishing accuracy, probably because the SecYEG dimer is a poor model of the engaged SecY (pdbs 5CH4 for engaged vs. 5AWW (Tanaka et al. 2015) and 2ZJS (Tsukazaki et al. 2008) for resting states). Another engaged structure proposes even greater squeezing of the “ring” when compared to the ellipse of the resting state, with simultaneous tightening of the lateral gate (pdb 5GAE (Jomaa et al. 2016)). How (a) substrate sampling between the aqueous environment of the channel and the hydrophobic membrane interior or (b) the translocation of co-translational substrates may occur under these conditions has remained unclear. The structures 3MP7 (Egea und Stroud 2010) and 5EUL (Li et al. 2016) are of high resolution and do show the expected ring widening and lateral gate opening. However, the former structure is that of the SecY dimer which raises the question of its relevance to the physiological event of protein translocation that now is believed to be conducted by the SecYEG monomer. The latter structure includes artificial cross-linking and a SecA-substrate fusion construct which may have biased the conformation.
Structural investigations are invaluable for understanding the intermolecular rearrangements during translocation. However, their interpretation is limited by several factors: First, stable translocation intermediates as stalled RNC (ribosome–nascent chain complexes) or proOmpA-SecA fusion products are all cross-linked to SecY [except for PDB 3J7R (Voorhees et al. 2014)]. Of course, cross-linking proves that the contact can take place, but it does not confirm its statistical relevance. Thus, in the absence of translocation dynamics we are lacking information as to whether the pdb snapshot reflects the prevalent translocon conformation during translocation.
Second, all high-resolution structures of SecY have been obtained either in detergent or in the cubic phase of monoacylglycerol monoolein. Both environments differ in terms of hydrophobic thickness, intrinsic curvature, surface charge, and pressure profile from the E. coli plasma membrane. Stretching forces at the lipid bilayer–water interfaces, compression in the membrane midplane (Cantor 1999) as well as electrostatic interactions or hydrophobic mismatch may well shift the equilibrium between the closed and open states. An example is provided by the interaction between negatively charged lipids and the N-terminus of SecA (Bauer et al. 2014). It forces SecA to be in a different conformation (Koch et al. 2016) than captured for the SecA-SecY structure in detergent (Zimmer et al. 2008).
Third, all of these structures lack transmembrane potential, to which SecY (but not Sec61) is exposed. In contrast to most of the structures with a translocation intermediate such as 3J46 (Park et al. 2013), 5GAE (Jomaa et al. 2016), and 5EUL (Li et al. 2016), the reconstituted SecY-signal peptide, SecY-proOmpA complex, or SecY-ribosome complex appear to be ion permeable at small transmembrane potentials (Knyazev et al. 2013; Knyazev et al. 2014). The complexes exclude ions when physiological values of membrane potential (Fig. 2b, d) are applied indicating that under physiological conditions, the conformation of the translocation intermediate is different from published structures (Knyazev et al. 2014). Assigning the functional properties of translocons at physiological temperatures to structural data obtained from translocons trapped in stable, low-energy conformations by cryo-EM or by X-ray crystallography is everything but straightforward. The potential problem of relating structural data obtained from low-energy conformations and functional data obtained at physiological temperatures is nothing peculiar to translocons, but it could be more important than it is for many other membrane proteins. For example, there has been excellent concordance between structural and functional data for K+ channels (Bezanilla 2008; Vargas et al. 2012). But, these channels have a rigid selectivity filter and relatively fixed conformations of inner and outer vestibules, with the exception of movements associated with gating. In contrast, it is clear from functional studies that the translocon has a very dynamic range of conformational states that can accommodate a large variety of protein structures during translocation. Therefore, it is very possible that the gating properties of the translocon could be dependent on structural rearrangements that will never appear in the stable structures obtained by cryo-EM or X-ray-crystallography. The importance of this point is underscored by the strong temperature dependence of the opening of the Sec61 translocon. Although sealing of the Sec61 pore by BiP was widely accepted, this sealing was electrophysiologically only observed at 5 °C (Wonderlin 2009), but the pore reopened with warming to physiological temperatures. From a biophysical point of view, the difference in temperature at which structural and functional data are collected is a potentially important stumbling block in reconciling different models for the structural basis for the functional properties of translocons.
Electrophysiological single channel measurements of SecYEG, purified and reconstituted into planar bilayers. a Single channel activity appeared only after addition of the signal peptide SP (N-terminal 20 residues from the pre-protein proOmpA). Numbers on the left correspond to the number of single channels, the distance between the two adjacent dashed lines corresponds to the current through a single channel. b An increase in transmembrane potential led to a decrease in membrane conductivity in a series of steps. Each conductivity step corresponds to the closure of a single channel. Conductivity almost fully recovered when no transmembrane potential was applied, indicating that SP remained bound to the lateral gate even in the closed state of SecY. c Experimental scheme for single SecYEG (green) channel measurements. The transmembrane potential was controlled via two AgCl-electrodes (grey) that were also used for monitoring bilayer (orange) conductivity. The bound proOmpA-DHFR is in blue. d Voltage-gated closing of SecYEG was also observed when the fusion protein proOmpA-DHFR was added instead of SP. The residual leakage is given in pA. e Putative scheme of voltage-driven channel closure. The resting channel is closed (left). SP binding opens the translocon (middle). It becomes permeable to the translocating peptide chain as well as to ions and water (represented by blue arrow) in the absence of physiological values of the transmembrane potential Δψ, which would close the channel (right). The stalled translocation intermediate is likely to be pushed from the lateral gate into the lipid. The upper panel shows the channel from the cytoplasm. TM stands for transmembrane helix. Single channel amplitude for A, B, D was 0.7 ± 0.1 pS/mM. Δψ is shown on the top. The small insets depict SecYEG (green) with a translocation intermediate (blue), SecYEG’s plug (red), and two of hydrophobic ring’s Ile (violet). The figure is taken from Knyazev et al. (2014). (Color figure online)
Maintenance of the Membrane Barrier to Small Molecules by SecY Engaged in Translocation as Reported by Electrophysiological and Other Functional Assays
Because the translocon is even closed to water in its resting state (Saparov et al. 2007), protein translocation requires a conformational transition to an open state. The lumen of the open state may be as large as 2.4 nm in diameter as has been observed with substrates that harboured rigid organic tails of different sizes (Bonardi et al. 2011). Such a large opening offers the possibility of translocating α helices in their folded state. In contrast, β-sheets have to pass in an unfolded state, since β barrels are even larger. Unfortunately, the resolution of available structures in the pdb databank is too low to unambiguously discern the secondary structure of α helical polypeptides in the SecY lumen (Table 1). However, there is little doubt that the signal sequence of pre-proteins is folded when bound in the lateral gate region (Fig. 1b). It was reported that folded α helical substrates are translocation-competent (Lecker et al. 1990), but they still might be unfolded during translocation. Unfolding may not be complete as successful translocation of proOmpA with an intramolecular loop formed by cross-linked cysteines was reported (Tani und Mizushima 1991).
Electrophysiological measurements of the purified and reconstituted SecYEG complex show SecYEG openings by pre-proteins or isolated signal peptides (Knyazev et al. 2014) (Fig. 2). Pore size estimates of ~ 0.4 nm from these measurements agree very well with those obtained by ion conductivity measurements of plug deletion mutants (Saparov et al. 2007). Such a diameter is large enough to allow unfolded peptide chains to be accommodated in the aqueous pore. It is truly surprising that pore size does not seem to depend on the presence of a peptide in the lateral gate (Knyazev et al. 2014), indicating that the translocation must possess unusual plasticity.
Ribosomes are also capable of opening the SecY channel (Knyazev et al. 2013), even though their affinity to the translocon seems to be smaller than that of pre-proteins. The same phenomenon has been reported for the eukaryotic translocon Sec61 (Erdmann et al. 2010), which may well result in an open state of the idle translocon at physiological temperatures (Wonderlin 2009). The resulting ion leak through the membrane of the endoplasmic reticulum was reported to be somehow inhibited by a calcium-dependent interaction of calmodulin with Sec61α (Erdmann et al. 2011).
Both prokaryotic and eukaryotic translocons appear to be voltage-gated. In both cases the probability that the channel was in the closed state increased with the absolute value of transmembrane potential (Knyazev et al. 2014; Wonderlin 2009; Erdmann et al. 2010). Since the membrane of the endoplasmic reticulum is thought to be de-energized, i.e. to possess no significant transmembrane potential, Sec61’s voltage dependence must be an evolutionary relict that the eukaryotic translocon inherited from its bacterial and archaeal ancestors. In contrast, voltage gating of SecY appears to be crucial for maintaining the membrane barrier to small cations and protons (Knyazev et al. 2014). The failure to do so would destroy the proton gradient across the bacterial plasma membrane, and thus would be lethal. The symmetric voltage dependence of translocon’s opening suggests that mobile sensor domains may not be the gating elements. Instead, voltage-driven bilayer thinning may gate the translocon. If so, electrostriction would induce a mismatch between the hydrophobic thicknesses of protein and membrane core. Mechanosensitive channels are known to respond to such a mismatch by a conformational change that eventually leads to channel opening (Perozo et al. 2002). Commonly a mechanical tension is applied that reduces membrane thickness by 2–5% (Sachs 2015). In contrast, electrostriction is less efficient. The bilayer thickness decreases with a factor \(2 \times {10^{ - 2}}/{V^2}\) (Alvarez und Latorre 1978). That is, for a transmembrane potential of Δψ ~ 100 mV we find \(\frac{{\Delta C}}{C}=\frac{{\Delta d}}{d}=\alpha {V^2}=2 \times {10^{ - 4}},\) or 0.02%, where C and d denote capacitance and thickness of the bilayer, respectively. Since hundredfold smaller that reported for the gating of mechanosensitive channels, this moderate thinning must be negligible. Otherwise, tiny temperature fluctuations would govern the translocon. This rules out electrostriction as a possible mechanism for gating the translocon.
Circumstantial evidence suggested that cation exclusion by SecY would help preserve the proton motive force (Dalal und Duong 2009). However, direct electrophysiological measurement revealed that SecY from E. coli prefers anions over cations by only about sevenfold (Sachelaru et al. 2017). The anionic selectivity of mammalian and yeast translocons from C. familiaris and S. cerevisiae is even smaller, with the permeability for anions being only about 1.5 times higher than for cations (Erdmann et al. 2010).
Role of the Plug
Interaction of the plug with the pore-forming helices is thought to stabilize the closed state of the SeYEG channel (Li et al. 2007). Accordingly, plug relocation from the constriction zone is likely to lead to lateral gate opening and widening of the hydrophobic ring. This is in line with cross-linking experiments in which immobilizing the plug in the vicinity of the SecE subunit (a forced open state) by a disulfide bridge renders SecY permeable to ions (Saparov et al. 2007). Disulfide bridge formation between these two rather distant elements in the crystal structure indicates that the plug must be rather flexible. Indeed, cross-linking of overexpressed SecY from inner membrane vesicles (IMVs) showed that plug movement can be restricted without abrogation of translocation. Therefore, the linker between the plug and helix 10 must be at least 8 Å in length (Lycklama a Nijeholt et al. 2010).
Contrary to the idea that plug dislocation is required for the opening of the lateral gate, the hydration of fluorescent labels on different positions on plug did not change upon insertion of co-translational translocation substrates (Lycklama a Nijeholt et al. 2010, 2011). Yet plug movement is accompanied by changes in the hydrophobicity of its environment as has been observed when the translocation intermediate consists of a post-translational substrate. To reconcile this observation with the ion channel activity that we see under comparable conditions for similarly de-energized reconstituted bilayers (Sachelaru et al. 2017), we have to assume that channel opening is not associated with plug movement but due to a widening of the pore ring.
Openings of the Lateral Gate
The opening of the lateral gate is essential for translocation. This follows from experiments with SecYEG containing IMVs, where bis-maleimides of different lengths were cross-linked to both poles of the lateral gate. Long linkers allowed translocation, whereas the shorter ones abrogated it (du Plessis et al. 2009).
Blocking both the eukaryotic Sec61α and the prokaryotic SecY in vivo by decatransin and cotransin (Junne et al. 2015; du Plessis et al. 2009) is in line with this conclusion. These blockers presumably bind to the constriction zone of the lateral gate in the closed state of the translocon. This way intercalation of the incoming signal peptide is prevented. Abrogating the tight ion seal of the closed translocon by introducing prl mutations (Saparov et al. 2007), also weakened inhibitor binding. This observation suggests that electrophysiological experiments on planar bilayers should be able to discern a smaller opening probability of the cotransin pre-treated translocon in the presence of pre-proteins.
ATP or GTP Hydrolysis as a Driving Force of Translocation
Co-translational translocation involves ribosome binding to the translocon and translocation is initiated while the elongation process of the translation is underway. The ribosome–SecY interaction interface is conserved and includes cytoplasmic loop 6/7 from the SecY side and ribosomal protein ul29 in prokaryotes (Prinz et al. 2000; Jomaa et al. 2016). The signal recognition particle (SRP) binds to a hydrophobic stretch of a signal sequence or transmembrane helix and helps to guide the nascent chain to the translocon with the help of the SRP-receptor (SR) in both eukaryotes and prokaryotes [for details see the review (Saraogi und Shan 2011)]. SRP-SR driven insertion of the first hydrophobic segment of the nascent chain into the translocon is thought to be GTP-dependent (Ataide et al. 2011). However, further translocation does not require SRP-SR (Miller und Walter 1993) and is conventionally assumed to be driven by the translational elongation, i.e. also GTP-driven. However, a polypeptide loop between the ribosome exit tunnel and the translocon [pdb 3J46, (Park et al. 2013)] rules out direct peptide pushing by the ribosome.
This is in line with the requirement for SecA to translocate the membrane protein RodZ, which has a 200 residues-long periplasmic domain (Wang et al. 2017). Obviously, bacterial ribosomal chain elongation per se does not provide a driving force for translocation of large cytoplasmic loops. It is worth noting that ribosomal chain elongation can still be rate limiting as alternative codons may lead to a threefold faster translocation (Sørensen und Pedersen 1991). Accordingly, by reducing the overall translational speed, the elongation inhibitor cycloheximide affects translocation, which in turn modifies the topology of membrane proteins (Goder und Spiess 2003).
In addition to GTP, the hydrophobic effect may fuel translocation: Once the elongating chain is hydrophobic enough, the free energy difference between its non-inserted and inserted states drives the chain into the lipid phase. The gain in energy may be large enough to overcome translation arrest (Ismail et al. 2012). For more hydrophilic substrates, the proton motive force may gain importance as an energy source for translocation. It has been shown to accelerate co-translational translocation (Ismail et al. 2015).
Post-translational translocation was proposed to be driven by the ATPase SecA (Hartl et al. 1990). Structural and biochemical research was applied to gain a more general understanding about the role of SecA homo-dimerization and binding kinetics to lipids and SecY. Under physiological conditions, SecA is predominantly a dimer with a cellular concentration of about 5 µM (Woodbury et al. 2002; Auclair et al. 2013). Within the translocon complex and upon lipid binding, SecA may act as a monomer (Or et al. 2005) or as a dimer (Kusters et al. 2011). That is, there is no consensus about the oligomeric state during translocation (Allen et al. 2016). The 20 N-terminal residues of SecA are important for binding to SecYEG and are also required for SecA-lipid interactions (Floyd et al. 2014; Bauer et al. 2014; Gouridis et al. 2013; Hendrick und Wickner 1991). Binding also depends on the nucleotide in SecA’s binding pocket: AMP-PNP, the non-hydrolysable analogue of ATP, augments SecA’s affinity to SecY by three orders of magnitude as compared to ADP (Deville et al. 2011).
Binding of the SecA-ATP complex may widen the SecY lumen while leaving the plug in a pore-sealing position (Zimmer et al. 2008; Allen et al. 2016) (Fig. 3b, pdb 3DIN in Table 1). However, the 3DIN structure has been captured in detergent and, thus, SecA’s amphipathic N-terminus adopts a position that is incompatible with its membrane interaction outlined above. For this interaction to occur, a major conformational change involving a 30 Å translational movement is required (Koch et al. 2016), which could potentially impact SecY’s plug position.
a Ribbon representation of the B. Subtilis SecA protein coloured by domain (Left) with the individual domains shown on the right (PDB 1M6N). They include (N-terminal to C-terminal on SecA): the nucleotide-binding domain-1 (NBD-1) (blue), the pre-protein cross-linking domain (PPXD) (gold), the nucleotide-binding domain-2 (NBD-2) (light blue), the central helix subdomain (CH) (green), the helical wing domain (HWD) (dark green), the two-helix finger subdomain (THF) (cyan), and the carboxyl-terminal linker (CTL). The CTL is depicted in red and serves as a model of PhoA signal peptide. From Zhang et al. (2017). b Cartoon of the SecA-SecYEG complex viewed from the side. Subunits of SecYEG and domains of SecA are subscribed according to colour code. The lines indicate the membrane boundaries. The two-helix finger of SecA inside the cytoplasmic funnel of SecY. From Zimmer et al. (2008). (Color figure online)
A scenario in which SecA regulates the state of SecY via ATP hydrolysis (Liang et al. 2009) is compatible with a mechanism in which SecA allows forward diffusion of the substrate, but blocks its backsliding. This hypothesis was also substantiated by single-molecule FRET experiments and molecular dynamics simulations (Allen et al. 2016). It is usually called Brownian ratcheting.
An alternative hypothesis of SecA-driven translocation envisions ATP hydrolysis to fuel conformational changes of SecA that result in an active pushing of the substrate. This power stroke hypothesis assigns the role of the pushing element to the two-helix finger (THF) of SecA (Zimmer et al. 2008; Bauer und Rapoport 2009) (Fig. 3a). SecA would push the bulky residues of the substrate, allowing the smaller ones to partially slide back (Bauer et al. 2014). This would allow the translocation of secretory proteins independent of their sequence—especially if the power stroke is coordinated with peptide binding and release from SecA’s pre-protein binding domain (PPXD).
However, it remains enigmatic how ATP hydrolysis in the NBD1 domain (Economou et al. 1995) translates into the big conformational change on the opposite side of the molecule, the THF domain, which is at the C-terminal part of the helical scaffold domain, HSD (for domain structure see (Hunt et al. 2002) and Fig. 3a). Thus, THF may have an alternative function. It may prime translocation by inserting the hairpin formed by the substrate’s signal peptide and the following segment (Zhang et al. 2017).
Translocation kinetics was explored in order to distinguish which model describes SecA-driven translocation best: power stroke or Brownian ratchet. The power stroke hypothesis for SecA was reported to be in line with the linear dependence of translocation time (τ) on the substrate length (N) in the absence of PMF (Tomkiewicz et al. 2006). Interestingly, such linear dependence τ(N) was subsequently reproduced in the presence of PMF but not in its absence (Liang et al. 2009) (Fig. 5b). This observation was interpreted in terms of a Brownian ratcheting mechanism in which SecA only functioned to regulate pore accessibility.
The dependency τ(N) was extensively researched for completely different translocation systems. For example, the translocation time of double-stranded DNA through solid state nanopores (made in alumina-coated silicon nitride) was found to be proportional to DNA length (Chen et al. 2004). The same was reported for single-stranded DNA in experiments with alpha hemolysin (αHL) pores (Kasianowicz et al. 1996). To ensure the polymer’s constant translocation velocity, the dragging force (here electrophoretic, acting on the negatively charged polymer when voltage was applied across the pore) must have been counterbalanced with the viscous force from the polymer–pore interaction.
In contrast to single- or double-stranded DNA that move through rigid pores, SecA-driven polypeptides may unfold on the cytoplasmic side or fold on the periplasmic side, thus delaying or accelerating translocation. Differences in the interaction of SecY’s translocation pore with various amino acid side chains only exacerbate the deviations from linearity. Such deviations were even found for the seemingly much more homogeneous DNA when passing through solid state nanopores (Storm et al. 2005). Thus, attributing a linear dependence τ(N) either to a power stroke or a Brownian ratcheting model does not appear to be straightforward.
Accessory Proteins May Drive Translocation
For a detailed review of SecY’s and Sec61’s translocation partners, the reader is referred elsewhere (Veenendaal et al. 2004; Park und Rapoport 2012; Lang et al. 2017). Here we only briefly discuss the topic from the perspective of accessory proteins as driving forces for translocation. Periplasmic chaperones such as Skp (Schäfer et al. 1999) or PpiD (Antonoaea et al. 2008) were reported to facilitate translocation via binding to the peptide chain which emerges from the periplasmic side. Thus, they prevent the polypeptide chain from backsliding, thereby contributing to the previously mentioned Brownian ratchet mechanism. Conceivably, the mitochondrial Hsp70 family chaperones operate according to a similar mechanism (Simon et al. 1992). In addition, to favour unidirectional diffusion, chaperones also assist in polypeptide folding. The resulting entropic pulling forces were reported to be above 10 pN (De Los Rios et al. 2006). The pulling comes with a penalty, due to a decrease in the chain’s entropy. This penalty diminishes with an increase in the total number of bound residues (Fig. 4).
Schematic view of translocation events that are driven by polypeptide folding (direction of translocation is from top to bottom). The number of possible conformations (in pink) without mtHsp70 is much larger than in its presence. That is, mtHsp70 binding to a translocating polypeptide reduces the number of “allowed” polypeptide conformations, i.e. conformations within the mitochondrial matrix space. It increases “forbidden” conformations (in white) due to additional spatial constraints. As a result, the accelerated folding process may exert a pulling force on the polypeptide within the translocon. From De Los Rios et al. (2006). (Color figure online)
Cross-linking experiments and co-purification of additional components alongside of SecYEG suggest there might be additional translocation interaction partners (Boy und Koch 2009; Sachelaru et al. 2014; Sachelaru et al. 2013). Of those, we will point out the proton channel SecDF which was reported as a possible component which couples the pH gradient and translocation (Arkowitz und Wickner 1994; Tsukazaki et al. 2011). In marine bacteria, the SecDF paralog exists and is suggested to employ the gradient of Na+ instead of H+ (Ishii et al. 2015). It was speculated that SecDF assists translocation by (i) first allowing its periplasmic P1 domain to bind to the emerging peptide chain, and (ii) second performing a conformational change that moves P1—along with the bound amino acids of the chain—away from the pore. SecDF is thought to use the proton gradient for both the conformation switch and binding-release cycles (Tsukazaki et al. 2011). This mechanism was supported by MD simulations (Ficici et al. 2017), which concluded that the P1 head movement is regulated by the transmembrane potential. The barrier for such transition is lowered in the presence of the transmembrane potential Δψ. The barrier for the reverse transition is lowered when Δψ is absent. Δψ oscillates along with SecDF conductivity, because proton flow along the existing pH gradient acts to reduce Δψ. This implies that substrate binding regulates SecDF’s open probability, and that SecDF is proton selective. The reported single channel amplitude of about 100 pS in neutral pH (Tsukazaki et al. 2011) is at odds with this anticipation. Since proton selective channels generally possess a tiny unitary conductivity that is too small to be resolved in single channel recordings (Decoursey 2003), the observation suggests that SecDF conducts other ions as well. SecDF is absent from Crenarchaea and organisms belonging to the thermoplasma group (at least from those which have been fully sequenced) (Bolhuis 2004). Many of those organisms are acidophiles, which might indicate that SecDF is not required in case of large transmembrane pH gradients.
It is worth mentioning that the minimal translocation machinery containing SecYEG is also capable of coupling Δψ to peptide translocation—even though with lower efficiency than the larger complex including SecDF (Schulze et al. 2014). A number of experiments lacking SecDF (Driessen und Wickner 1991; Liang et al. 2009) show a Δψ -mediated acceleration of the translocation (Fig. 5a, b). Thus, SecDF does not appear to be essential for translocation, but it may facilitate translocation by a yet unknown mechanism.
a Rate of translocation is stimulated by PMF (Eq. 1). Δψ and ΔpH are interchangeable. The figure is taken from Driessen und Wickner (1991). b In the presence of PMF, the time required for translocation (= transport time) increases linearly with the length of the polypeptide (= precursor length) that is translocated. The figure is taken from Liang et al. (2009). c PMF acts by exerting an electric pulling force on nascent chains containing negatively charged residues (located in the region marked in red). The force is sufficient to trigger the release from translation arrest (arrest peptide in blue) that has been imposed by SecM. fFL stands for the fraction of the peptide that has been translocated. The x axis shows the distance from the arrest peptide to the charged stretch (red). fFL was largest when the red segment contained acidic residues (5D). However, it also increased in case of basic (5K) and non-charged (5N, 5Q) residues. Simple pulling by Δψ cannot explain this observation. The panel is taken from Ismail et al. (2015). The colour schemes on top describe the experimental system used: in vitro post-translational translocation for A–B and co-translational translocation for C. (Color figure online)
PMF-Driven Translocation
SecYEG resides in a permanently energized membrane—the cell sustains the electrochemical gradient of protons across the cytoplasmic membrane, also known as PMF. This energy might be utilized by the translocation system, although probably not by the eukaryotic one, as no significant PMF is built across the membrane of the endoplasmic reticulum.
PMF consists of electrical (transmembrane potential Δψ) and chemical (pH gradient) components:
where R and T represent the gas constant and absolute temperature, respectively.
The first observation of the translocation rate increasing effect of the electrochemical gradient of protons was made on inner membrane vesicles containing SecYE (Driessen und Wickner 1991) (Fig. 5a). The proton–potassium antiporter nigericin was used to dissipate ΔpH, the potassium ionophore valinomycin—to dissipate Δψ. The changes in inner pH and transmembrane potential were monitored with fluorescent sensors pyranine and Oxonol VI. PMF seemed to accelerate transport, even though Δψ across the bilayer of IMVs was smaller than in E. coli cells: judging from Oxonol VI quenching (~ 10%), the absolute value of Δψ was only about 10 mV. This estimate is much smaller than what is usually expected for IMVs: Δψ ~ 80 mV (Keyzer 2002). The reason for the discrepancy is unclear. Proton pumping and proton consumption by SecY may have anyhow resulted in a non-uniform Δψ distribution along the IMV bilayer. Subsequent measurements with a fluorescence translocation assay (Liang et al. 2009) also showed that PMF increases the translocation rate. Moreover, complete translocation of proOmpA with an intramolecular crosslink between residues 290 and 302 was only observed in the presence of PMF, suggesting that PMF is essential for the translocation of larger substrates (Tani und Mizushima 1991).
How does each of the two PMF components facilitate translocation? The SecM stalling sequence was used in vivo as a force probe to estimate the pulling force that Δψ exerts on the nascent chain during translocation (Fig. 5c) (Ismail et al. 2015). Δψ stimulates movement of acidic residues through the translocon, thereby releasing the translation arrest of the nascent chain. Conceivably, Δψ was not the sole driving force as segments enriched in basic residues were also freed from the arrest.
Δψ does not impose the “positive inside” rule by retarding the passage of positive charges, i.e. it is not responsible for the topology of membrane proteins. Otherwise the reversal of Δψ in acidophiles would have led to an enrichment of the periplasmic leaflet with the positively charged amino acid residues arginine and lysine. Rather anionic lipids act to anchor the positive charges to the cytosolic leaflet enabling acidophiles to follow the positive inside rule (van de Vossenberg et al. 1998).
According to (Zilberstein et al. 1979) PMF in living E. coli cell amounts to 150–200 mV. With Δψ ranging from 100 to 150 mV (negative in cytoplasm), trivial electrostatic considerations give a pulling force that acts on a single elementary negative charge of about:
which is comparable to the force developed by Hsp70-assisted folding (see previous section) or by unassisted nascent chain folding, which develops forces in the range from 4 to 8 pN (Schlierf et al. 2007).
Proteins are equally enriched in acidic and basic residues. Δψ only pulls acidic residues from the cytosol into the periplasmic space, but it moves basic residues in the opposite direction. Nevertheless, the accelerating effect of PMF on translocation (Daniels et al. 1981; Yamane et al. 1987) can—although to a lower degree—also be observed for substrates enriched in basic residues (Liang et al. 2012). The decline in rate and efficiency of translocation both depended on the distribution of basic residues and their total number. Thus, the positive PMF effect on the translocation of stretches with positively charged amino acid side chains indicates that ΔpH must be involved. This conclusion is in line with the observation that the translocation rate dropped to 30% in heavy water. Such an isotope effect can only be explained by the involvement of proton transfer reactions (Springer et al. 2011). ΔpH was reported to somehow facilitate the passage of (i) bulkier substrates (Bonardi et al. 2011) or (ii) larger polypeptide segments after their release from SecA (Mitra et al. 2006). For translocation of other substrates, ΔpH does not seem to be essential (Koch und Müller 2000).
Currently it is not understood how ΔpH drives translocation. Any putative mechanism should account (i) for the lack of a net charge since acidic and basic residues are roughly equal in number for most polypeptides and (ii) for the variability of ΔpH and Δψ in neutralo-, acido-, and alkali-philes. E. coli belongs to the neutralophiles and hence, it has a moderate ΔpH of about 0.5–1.5 pH units (acidic in the periplasm) and a high Δψ of about − 100 to − 150 mV (negative in the cytoplasm) (Krulwich et al. 2011). Acidophiles have a larger ΔpH but a smaller Δψ of opposite polarity (Ingledew 1990). Alkaliphiles have a moderate ΔpH (basic in periplasm), but a large Δψ (negative in cytoplasm) (Sturr et al. 1994). We believe that the considerable pKa shift of titratable residues within the translocon is key for the understanding of how a universal mechanism may cope with such diversity.
The Putative Mechanism of PMF-Driven Translocation
Both pKa shifts and a pH microclimate within the translocon appear to be crucial to the PMF-driven translocation.
-
(i)
The pKa values of both basic and acidic residues shift towards neutral values within the translocation pore due to the well-known dependence of pKa on the dielectric properties of the environment. It is thought to be based on minimizing the electrostatic energy penalty known as Born energy for putting a charge into a medium with low electric permittivity. The relative permittivity εr amounts to ~ 2 in the lipid bilayer core (Huang und Levitt 1977) and to ~ 3 inside proteins (Kukic et al. 2013). The low εr value agrees with the observed high ordering of water molecules inside the translocon by molecular dynamic simulations (Capponi et al. 2015). A pKa shift of 3–5 pH units was reported for pH-sensitive dyes in dioxane-water mixtures upon lowering the dielectric constant εr from 80 to 10 (Fernandez und Fromherz 1977). Similarly, pKa shifts of up to 5 pH units were observed for both acidic (Isom et al. 2010) and basic residues (Isom et al. 2011; Robinson et al. 2014), when transferring them into apolar proteinaceous surroundings.
-
(ii)
pH within the translocon pore is more basic than in the cytoplasm. This is due to SecY’s moderate anionic selectivity: Since the permeability for anions is sevenfold higher than for cations (Sachelaru et al. 2017), positively charged particles must be partially excluded from the pore. Under physiological PMF this pH shift is expected to be even larger, because voltage gating of SecY (Knyazev et al. 2014) narrows the translocation pore, which in turn exacerbates electrostatic effects.
To reflect the heterogeneity of ε in the membrane, we divide the substrate pathway into three slabs. In the outer two (light blue on Fig. 6), we assume ε to be close enough to its bulk value so that the titratable residues will have bulk-like pKa values. In the middle slab (green), pKa will be shifted due its low ε.
PMF-driven translocation. Both pH microclimate and pKa shifts in the constriction zone of the translocon (grey), which alter the probability of being charged for both basic (blue) and acidic residues (red) to the effect that only the forward movement (towards the periplasm) of the translocating peptide chain (black line) is facilitated by the transmembrane potential Δψ (white arrows). Left panel: Slabs with three different dielectric permittivities are distinguished and the resulting approximate pKa of titratable amino acid side chains is indicated for both acidic (red) and basic (blue) residues. Right panels: Indicates whether the titratable amino acid side chains are expected to be charged (plus or minus inside a circle) or uncharged (empty circles). ΔpH values and Δψ polarities are taken from literature: (Krulwich et al. 2011; Zilberstein et al. 1979; Ingledew 1990; Sturr et al. 1994), left to right. (Color figure online)
Since the channel has an hourglass shape (Fig. 1), the middle slab is also the pore restriction site for secretory proteins. One would obviously expect channel resistivity to ion flow be greatest at this most constricted region of the pore. As a result, the electrical transmembrane field is focussed onto exactly the same spot where the shift in ε is largest. Local charged groups or the surface charge on protruding lipids may affect the dielectric properties of translocon’s pore, but they do not shift the constriction zone. That is, the region of steepest potential profile remains well aligned with the region in which pKa is most affected.
Consequently, we may assume the pKa of basic residues (lysine and arginine) to be equal to about 11 in the outer slabs, and that of acidic residues (glutamate and aspartate) to be equal to about 4. These pKa values shift to about 7.5 for both types of residues in the middle slab. pH within the two outer slabs is equal to the respective bulk values of cytoplasm and periplasm. pH of the middle slab is equal to their arithmetic mean to which the pH shift \(\delta (pH)\) has to be added which arises from translocon’s anion selectivity. Based on these assumptions we arrive at pH ≈ 8 for the middle slab (Fig. 6, Neutralophile).
The pH microclimate in the constriction zone of the translocon renders the probabilities of being charged unequal for basic and acidic residues. In consequence, Δψ ensures the uni-directionality of polypeptide translocation from the cytoplasmic to periplasmic sides (Fig. 6). Specifically, the acidic residues of amino acid side chains are likely to be charged when the polypeptide traverses the constriction side of the translocon in neutralophiles (E. coli) and alkaliphiles (Bacillus pseudofirmus). Consequently, Δψ exerts a net electrostatic force on the peptide chain that is directed towards the periplasm (Fig. 6). In contrast, polypeptide’s basic residues are charged for acidophiles (Acidithiobacillus ferrooxidans). The net electrostatic force will still be directed towards the periplasm since Δψ has an inverted polarity in acidophiles (Fig. 6).
To prove the proposed mechanism of PMF-driven translocation (Fig. 6), precise control over both ΔpH and Δψ must be executed in the experimental system. This requirement rules out both IMVs and reconstituted vesicles. From that perspective, SecY reconstitution into free-spanning lipid bilayers appears most appealing—if combined with single-molecule fluorescence techniques.
References
Allen WJ, Corey RA, Oatley P, Sessions RB, Radford SE, Tuma R, Collinson I (2016) Two-way communication between SecY and SecA suggests a Brownian ratchet mechanism for protein translocation. eLife. https://doi.org/10.7554/eLife.15598
Alvarez O, Latorre R (1978) Voltage-dependent capacitance in lipid bilayers made from monolayers. Biophys J 21(1):1–17. https://doi.org/10.1016/S0006-3495(78)85505-2
Antonoaea R, Fürst M, Nishiyama K-I, Müller M (2008) The periplasmic chaperone PpiD interacts with secretory proteins exiting from the SecYEG translocon. Biochemistry 47(20):5649–5656. https://doi.org/10.1021/bi800233w
Arkowitz RA, Wickner W (1994) SecD and SecF are required for the proton electrochemical gradient stimulation of preprotein translocation. EMBO J 13(4):954–963
Ataide SF, Schmitz N, Shen K, Ke A, Shan S-o, Doudna JA, Ban N (2011) The crystal structure of the signal recognition particle in complex with its receptor. Science (New York) 331(6019):881–886. https://doi.org/10.1126/science.1196473
Auclair SM, Oliver DB, Mukerji I (2013) Defining the solution state dimer structure of Escherichia coli SecA using Förster resonance energy transfer. Biochemistry 52(14):2388–2401. https://doi.org/10.1021/bi301217t
Bauer BW, Rapoport TA (2009) Mapping polypeptide interactions of the SecA ATPase during translocation. Proc Natl Acad Sci USA 106(49):S20800–S20805. https://doi.org/10.1073/pnas.0910550106
Bauer BW, Shemesh T, Chen Yu, Rapoport TA (2014) A “push and slide” mechanism allows sequence-insensitive translocation of secretory proteins by the SecA ATPase. Cell 157(6):1416–1429. https://doi.org/10.1016/j.cell.2014.03.063
Bezanilla F (2008) How membrane proteins sense voltage. Nat Rev Mol Cell Biol 9(4):323–332. https://doi.org/10.1038/nrm2376
Bolhuis A (2004) The archaeal Sec-dependent protein translocation pathway. Philos Trans R Soc B 359(1446):919–927. https://doi.org/10.1098/rstb.2003.1461
Bonardi F, Halza E, Walko M, Du Plessis F, Nouwen N, Feringa BL, Driessen AJM (2011) Probing the SecYEG translocation pore size with preproteins conjugated with sizable rigid spherical molecules. Proc Natl Acad Sci USA 108(19):S7775–s7780. https://doi.org/10.1073/pnas.1101705108
Bondar A-N, del Val C, Freites JA, Tobias DJ, White SH (2010) Dynamics of SecY translocons with translocation-defective mutations. Structure 18(7):847–857. https://doi.org/10.1016/j.str.2010.04.010
Boy D, Koch H-G (2009) Visualization of distinct entities of the SecYEG translocon during translocation and integration of bacterial proteins. Mol Biol Cell 20(6):1804–1815. https://doi.org/10.1091/mbc.E08-08-0886
Cantor RS (1999) Lipid composition and the lateral pressure profile in bilayers. Biophys J 76(5):2625–2639. https://doi.org/10.1016/S0006-3495(99)77415-1
Capponi S, Heyden M, Bondar A-N, Tobias DJ, White SH (2015) Anomalous behavior of water inside the SecY translocon. Proc Natl Acad Sci USA 112(29):9016–9021. https://doi.org/10.1073/pnas.1424483112
Chen P, Gu J, Brandin E, Kim Y-R, Wang Q, Branton D (2004) Probing single DNA molecule transport using fabricated nanopores. Nano Lett 4(11):2293–2298. https://doi.org/10.1021/nl048654j
Dalal K, Duong F (2009) The SecY complex forms a channel capable of ionic discrimination. EMBO Rep 10(7):762–768. https://doi.org/10.1038/embor.2009.87
Daniels CJ, Bole DG, Quay SC, Oxender DL (1981) Role for membrane potential in the secretion of protein into the periplasm of Escherichia coli. Proc Natl Acad Sci USA 78(9):S5396–S5400
De Los Rios P, Ben-Zvi A, Slutsky O, Azem A, Goloubinoff P (2006) Hsp70 chaperones accelerate protein translocation and the unfolding of stable protein aggregates by entropic pulling. Proc Natl Acad Sci USA 103(16):S6166–S6171. https://doi.org/10.1073/pnas.0510496103
Decoursey TE (2003) Voltage-gated proton channels and other proton transfer pathways. Physiol Rev 83(2):475–579. https://doi.org/10.1152/physrev.00028.2002
Denks K, Vogt A, Sachelaru I, Petriman N-A, Kudva R, Koch H-G (2014) The Sec translocon mediated protein transport in prokaryotes and eukaryotes. Mol Membr Biol 31(2–3):58–84. https://doi.org/10.3109/09687688.2014.907455
Deville K, Gold VAM, Robson A, Whitehouse S, Sessions RB, Baldwin SA et al (2011) The oligomeric state and arrangement of the active bacterial translocon. J Biol Chem 286(6):4659–4669. https://doi.org/10.1074/jbc.M110.175638
Driessen AJ, Wickner W (1991) Proton transfer is rate-limiting for translocation of precursor proteins by the Escherichia coli translocase. Proc Natl Acad Sci USA 88(6):S2471–S2475
du Plessis DJF, Berrelkamp G, Nouwen N, Driessen AJM (2009) The lateral gate of SecYEG opens during protein translocation. J Biol Chem 284(23):15805–15814. https://doi.org/10.1074/jbc.M901855200
Economou A, Pogliano JA, Beckwith J, Oliver DB, Wickner W (1995) SecA membrane cycling at SecYEG is driven by distinct ATP binding and hydrolysis events and is regulated by SecD and SecF. Cell 83(7):1171–1181. https://doi.org/10.1016/0092-8674(95)90143-4
Egea PF, Stroud RM (2010) Lateral opening of a translocon upon entry of protein suggests the mechanism of insertion into membranes. Proc Natl Acad Sci USA 107(40):S17182–S17187. https://doi.org/10.1073/pnas.1012556107
Erdmann F, Jung M, Maurer P, Harsman A, Zimmermann R, Wagner R (2010) The mammalian and yeast translocon complexes comprise a characteristic Sec61 channel. Biochem Biophys Res Commun 396(3):714–720. https://doi.org/10.1016/j.bbrc.2010.04.168
Erdmann F, Schäuble N, Lang S, Jung M, Honigmann A, Ahmad M et al (2011) Interaction of calmodulin with Sec61α limits Ca2+ leakage from the endoplasmic reticulum. EMBO J 30(1):17–31. https://doi.org/10.1038/emboj.2010.284
Fernandez MS, Fromherz P (1977) Lipoid pH indicators as probes of electrical potential and polarity in micelles. J Phys Chem 81(18):1755–1761. https://doi.org/10.1021/j100533a009
Ficici E, Jeong D, Andricioaei I (2017) Electric-field-induced protein translocation via a conformational transition in SecDF. An MD study. Biophys J 112(12):2520–2528. https://doi.org/10.1016/j.bpj.2017.04.034
Floyd JH, You Z, Hsieh Y-H, Ma Y, Yang H, Tai PC (2014) The dispensability and requirement of SecA N-terminal aminoacyl residues for complementation, membrane binding, lipid-specific domains and channel activities. Biochem Biophys Res Commun 453(1):138–142. https://doi.org/10.1016/j.bbrc.2014.09.080
Goder V, Spiess M (2003) Molecular mechanism of signal sequence orientation in the endoplasmic reticulum. EMBO J 22(14):3645–3653. https://doi.org/10.1093/emboj/cdg361
Gouridis G, Karamanou S, Sardis MF, Scharer MA, Capitani G, Economou A (2013) Quaternary dynamics of the SecA motor drive translocase catalysis. Mol Cell 52(5):655–666. https://doi.org/10.1016/j.molcel.2013.10.036
Hartl FU, Lecker S, Schiebel E, Hendrick JP, Wickner W (1990) The binding cascade of SecB to SecA to SecY/E mediates preprotein targeting to the E. coli plasma membrane. Cell 63(2):269–279
Hendrick JP, Wickner W (1991) SecA protein needs both acidic phospholipids and SecY/E protein for functional high-affinity binding to the Escherichia coli plasma membrane. J Biol Chem 266(36):24596–24600
Huang W, Levitt DG (1977) Theoretical calculation of the dielectric constant of a bilayer membrane. Biophys J 17(2):111–128. https://doi.org/10.1016/S0006-3495(77)85630-0
Huber D, Rajagopalan N, Preissler S, Rocco MA, Merz F, Kramer G, Bukau B (2011) SecA interacts with ribosomes in order to facilitate posttranslational translocation in bacteria. Mol Cell 41(3):343–353. https://doi.org/10.1016/j.molcel.2010.12.028
Hunt JF, Weinkauf S, Henry L, Fak JJ, McNicholas P, Oliver DB, Deisenhofer J (2002) Nucleotide control of interdomain interactions in the conformational reaction cycle of SecA. Science (New York) 297(5589):2018–2026. https://doi.org/10.1126/science.1074424
Ingledew WJ (1990) Acidophiles. In: Edwards C (ed) Microbiology of extreme environments. McGraw-Hill, pp 33–54
Ishii E, Chiba S, Hashimoto N, Kojima S, Homma M, Ito K et al (2015) Nascent chain-monitored remodeling of the Sec machinery for salinity adaptation of marine bacteria. Proc Natl Acad Sci USA 112(40):E5513–E5522. https://doi.org/10.1073/pnas.1513001112
Ismail N, Hedman R, Lindén M, von Heijne G (2015) Charge-driven dynamics of nascent-chain movement through the SecYEG translocon. Nat Struct Mol Biol 22(2):145–149. https://doi.org/10.1038/nsmb.2940
Ismail N, Hedman R, Schiller N, von Heijne G (2012) A biphasic pulling force acts on transmembrane helices during translocon-mediated membrane integration. Nat Struct Mol Biol 19(10):1018–1022. https://doi.org/10.1038/nsmb.2376
Isom DG, Castaneda CA, Cannon BR, Garcia-Moreno EB (2011) Large shifts in pKa values of lysine residues buried inside a protein. Proc Natl Acad Sci USA 108(13):S5260–S5265. https://doi.org/10.1073/pnas.1010750108
Isom DG, Castañeda CA, Cannon BR, Velu PD, García-Moreno E, Bertrand (2010) Charges in the hydrophobic interior of proteins. Proc Natl Acad Sci USA 107(37):S16096–S16100. https://doi.org/10.1073/pnas.1004213107
Jomaa A, Boehringer D, Leibundgut M, Ban N (2016) Structures of the E. coli translating ribosome with SRP and its receptor and with the translocon. Nat Comms 7:10471. https://doi.org/10.1038/ncomms10471
Junne T, Schwede T, Goder V, Spiess M (2007) Mutations in the Sec61p channel affecting signal sequence recognition and membrane protein topology. J Biol Chem 282(45):33201–33209. https://doi.org/10.1074/jbc.M707219200
Junne T, Wong J, Studer C, Aust T, Bauer BW, Beibel M et al (2015) Decatransin, a new natural product inhibiting protein translocation at the Sec61/SecYEG translocon. J Cell Sci 128(6):1217–1229. https://doi.org/10.1242/jcs.165746
Kasianowicz JJ, Brandin E, Branton D, Deamer DW (1996) Characterization of individual polynucleotide molecules using a membrane channel. Proc Natl Acad Sci USA 93(24):13770–13773
Keyzer J de (2002) Kinetic analysis of the translocation of fluorescent precursor proteins into Escherichia coli membrane vesicles. J Biol Chem 277(48):46059–46065. https://doi.org/10.1074/jbc.M208449200
Knyazev DG, Lents A, Krause E, Ollinger N, Siligan C, Papinski D et al (2013) The bacterial translocon SecYEG opens upon ribosome binding. J Biol Chem 288(25):17941–17946. https://doi.org/10.1074/jbc.M113.477893
Knyazev DG, Winter L, Bauer BW, Siligan C, Pohl P (2014) Ion conductivity of the bacterial translocation channel SecYEG engaged in translocation. J Biol Chem 289(35):24611–24616. https://doi.org/10.1074/jbc.M114.588491
Koch HG, Müller M (2000) Dissecting the translocase and integrase functions of the Escherichia coli SecYEG translocon. J Cell Biol 150(3):689–694
Koch S, Wit JG, de Vos I, Birkner JP, Gordiichuk P, Herrmann A et al (2016) Lipids activate SecA for high affinity binding to the SecYEG complex. J Biol Chem 291(43):22534–22543. https://doi.org/10.1074/jbc.M116.743831
Krulwich TA, Sachs G, Padan E (2011) Molecular aspects of bacterial pH sensing and homeostasis. Nat Rev Microbiol 9(5):330–343. https://doi.org/10.1038/nrmicro2549
Kuhn P, Weiche B, Sturm L, Sommer E, Drepper F, Warscheid B et al (2011) The bacterial SRP receptor, SecA and the ribosome use overlapping binding sites on the SecY translocon. Traffic 12(5):563–578. https://doi.org/10.1111/j.1600-0854.2011.01167.x
Kukic P, Farrell D, McIntosh LP, García-Moreno E, Jensen B, Toleikis KS, Zigmantas et al (2013) Protein dielectric constants determined from NMR chemical shift perturbations. J Am Chem Soc 135(45):16968–16976
Kusters I, van den Bogaart G, Kedrov A, Krasnikov V, Fulyani F, Poolman B, Driessen AJM (2011) Quaternary structure of SecA in solution and bound to SecYEG probed at the single molecule level. Structure 19(3):430–439. https://doi.org/10.1016/j.str.2010.12.016
Lang S, Pfeffer S, Lee P-H, Cavalié A, Helms V, Förster F, Zimmermann R (2017) An update on Sec61 channel functions, mechanisms, and related diseases. Front Physiol 8:225. https://doi.org/10.3389/fphys.2017.00887
Lecker SH, Driessen AJ, Wickner W (1990) ProOmpA contains secondary and tertiary structure prior to translocation and is shielded from aggregation by association with SecB protein. EMBO J 9(7):2309–2314
Li L, Park E, Ling JJ, Ingram J, Ploegh H, Rapoport TA (2016) Crystal structure of a substrate-engaged SecY protein-translocation channel. Nature 531(7594):395–399. https://doi.org/10.1038/nature17163
Li W, Schulman S, Boyd D, Erlandson K, Beckwith J, Rapoport TA (2007) The plug domain of the SecY protein stabilizes the closed state of the translocation channel and maintains a membrane seal. Mol Cell 26(4):511–521. https://doi.org/10.1016/j.molcel.2007.05.002
Liang F-C, Bageshwar UK, Musser SM (2009) Bacterial Sec protein transport is rate-limited by precursor length: a single turnover study. Mol Biol Cell 20(19):4256–4266. https://doi.org/10.1091/mbc.E09-01-0075
Liang F-C, Bageshwar UK, Musser SM (2012) Position-dependent effects of polylysine on Sec protein transport. J Biol Chem 287(16):12703–12714. https://doi.org/10.1074/jbc.M111.240903
Lycklama a Nijeholt JA, Bulacu M, Marrink SJ, Driessen AJM (2010) Immobilization of the plug domain inside the SecY channel allows unrestricted protein translocation. J Biol Chem 285(31):23747–23754. https://doi.org/10.1074/jbc.M110.124636
Lycklama a Nijeholt JA, Wu ZC, Driessen AJM (2011) Conformational Dynamics of the plug domain of the SecYEG protein-conducting channel. J Biol Chem 286(51):43881–43890. https://doi.org/10.1074/jbc.M111.297507
Miller JD, Walter P (1993) A GTPase cycle in initiation of protein translocation across the endoplasmic reticulum membrane. Ciba Found Sympos 176:147–159 (discussion 159–163)
Mitra K, Frank J, Driessen A (2006) Co- and post-translational translocation through the protein-conducting channel: analogous mechanisms at work? Nat Struct Mol Biol 13(11):957–964. https://doi.org/10.1038/nsmb1166
Or E, Boyd D, Gon S, Beckwith J, Rapoport T (2005) The bacterial ATPase SecA functions as a monomer in protein translocation. J Biol Chem 280(10):9097–9105. https://doi.org/10.1074/jbc.M413947200
Park E, Ménétret J-F, Gumbart JC, Ludtke SJ, Li W, Whynot A et al (2013) Structure of the SecY channel during initiation of protein translocation. Nature 506(7486):102–106. https://doi.org/10.1038/nature12720
Park E, Rapoport TA (2012) Mechanisms of Sec61/SecY-mediated protein translocation across membranes. Ann Rev Biophys 41(1):21–40. https://doi.org/10.1146/annurev-biophys-050511-102312
Perozo E, Cortes D, Sompornpisut P, Kloda A, Martinac B (2002) Open channel structure of MscL and the gating mechanism of mechanosensitive channels. Nature 418(6901):942–948. https://doi.org/10.1038/nature00992
Prinz A, Behrens C, Rapoport TA, Hartmann E, Kalies KU (2000) Evolutionarily conserved binding of ribosomes to the translocation channel via the large ribosomal RNA. EMBO J 19(8):1900–1906. https://doi.org/10.1093/emboj/19.8.1900
Robinson AC, Castaneda CA, Schlessman JL, Garcia-Moreno E, Bertrand (2014) Structural and thermodynamic consequences of burial of an artificial ion pair in the hydrophobic interior of a protein. Proc Natl Acad Sci USA 111(32):S11685–S11690. https://doi.org/10.1073/pnas.1402900111
Sachelaru I, Petriman NA, Kudva R, Kuhn P, Welte T, Knapp B et al (2013) YidC occupies the lateral gate of the SecYEG translocon and is sequentially displaced by a nascent membrane protein. J Biol Chem 288(23):16295–16307. https://doi.org/10.1074/jbc.M112.446583
Sachelaru I, Petriman N-A, Kudva R, Koch H-G (2014) Dynamic interaction of the sec translocon with the chaperone PpiD. J Biol Chem 289(31):21706–21715. https://doi.org/10.1074/jbc.M114.577916
Sachelaru I, Winter L, Knyazev DG, Zimmermann M, Vogt A, Kuttner R et al (2017) YidC and SecYEG form a heterotetrameric protein translocation channel. Sci Rep 7(1):101. https://doi.org/10.1038/s41598-017-00109-8
Sachs F (2015) Mechanical transduction by ion channels. A cautionary tale. World J Neurol 5(3):74–87. https://doi.org/10.5316/wjn.v5.i3.74
Saparov SM, Erlandson K, Cannon K, Schaletzky J, Schulman S, Rapoport TA, Pohl P (2007) Determining the conductance of the SecY protein translocation channel for small molecules. Mol Cell 26(4):501–509. https://doi.org/10.1016/j.molcel.2007.03.022
Saraogi I, Shan S-o (2011) Molecular mechanism of co-translational protein targeting by the signal recognition particle. Traffic 12(5):535–542. https://doi.org/10.1111/j.1600-0854.2011.01171.x
Schäfer U, Beck K, Müller M (1999) Skp, a molecular chaperone of gram-negative bacteria, is required for the formation of soluble periplasmic intermediates of outer membrane proteins. J Biol Chem 274(35):24567–24574
Schlierf M, Berkemeier F, Rief M (2007) Direct observation of active protein folding using lock-in force spectroscopy. Biophys J 93(11):3989–3998. https://doi.org/10.1529/biophysj.107.114397
Schulze RJ, Komar J, Botte M, Allen WJ, Whitehouse S, Gold VAM et al (2014) Membrane protein insertion and proton-motive-force-dependent secretion through the bacterial holo-translocon SecYEG–SecDF–YajC–YidC. Proc Natl Acad Sci USA 111(13):4844–4849. https://doi.org/10.1073/pnas.1315901111
Simon SM, Blobel G (1992) Signal peptides open protein-conducting channels in E. coli. Cell 69(4):677–684
Simon SM, Peskin CS, Oster GF (1992) What drives the translocation of proteins? Proc Natl Acad Sci USA 89(9):S3770–S3774
Sørensen MA, Pedersen S (1991) Absolute in vivo translation rates of individual codons in Escherichia coli. The two glutamic acid codons GAA and GAG are translated with a threefold difference in rate. J Mol Biol 222(2):265–280
Springer A, Hagen V, Cherepanov DA, Antonenko YN, Pohl P (2011) Protons migrate along interfacial water without significant contributions from jumps between ionizable groups on the membrane surface. Proc Natl Acad Sci USA 108(35):S14461–S14466. https://doi.org/10.1073/pnas.1107476108
Storm AJ, Chen JH, Zandbergen HW, Dekker C (2005) Translocation of double-strand DNA through a silicon oxide nanopore. Phys Rev E 71(5 Pt 1):51903. https://doi.org/10.1103/PhysRevE.71.051903
Sturr MG, Guffanti AA, Krulwich TA (1994) Growth and bioenergetics of alkaliphilic Bacillus firmus OF4 in continuous culture at high pH. J Bacteriol 176(11):3111–3116
Tanaka Y, Sugano Y, Takemoto M, Mori T, Furukawa A, Kusakizako T et al (2015) Crystal structures of SecYEG in lipidic cubic phase elucidate a precise resting and a peptide-bound state. Cell Rep 13(8):1561–1568. https://doi.org/10.1016/j.celrep.2015.10.025
Tani K, Mizushima S (1991) A chemically cross-linked nonlinear proOmpA molecule can be translocated into everted membrane vesicles of Escherichia coli in the presence of the proton motive force. FEBS Lett 285(1):127–131
Tomkiewicz D, Nouwen N, van Leeuwen R, Tans S, Driessen AJM (2006) SecA supports a constant rate of preprotein translocation. J Biol Chem 281(23):15709–15713. https://doi.org/10.1074/jbc.M600205200
Tsukazaki T, Mori H, Echizen Y, Ishitani R, Fukai S, Tanaka T et al (2011) Structure and function of a membrane component SecDF that enhances protein export. Nature 474(7350):235–238. https://doi.org/10.1038/nature09980
Tsukazaki T, Mori H, Fukai S, Ishitani R, Mori T, Dohmae N et al (2008) Conformational transition of Sec machinery inferred from bacterial SecYE structures. Nature 455(7215):988–991. https://doi.org/10.1038/nature07421
van de Vossenberg JLCM, Albers S-V, van der Does C, Driessen AJM, van Klompenburg W (1998) The positive inside rule is not determined by the polarity of the ∆ψ. Mol Microbiol 29(4):1125–1126. https://doi.org/10.1046/j.1365-2958.1998.01001.x
van den Berg B, Clemons WM, Collinson I, Modis Y, Hartmann E, Harrison SC, Rapoport TA (2004) X-ray structure of a protein-conducting channel. Nature 427(6969):36–44. https://doi.org/10.1038/nature02218
Vargas E, Yarov-Yarovoy V, Khalili-Araghi F, Catterall WA, Klein ML, Tarek M et al (2012) An emerging consensus on voltage-dependent gating from computational modeling and molecular dynamics simulations. J Gen Physiol 140(6):587–594. https://doi.org/10.1085/jgp.201210873
Veenendaal AK, van der Does C, Driessen AJ (2004) The protein-conducting channel SecYEG. Biochim Biophys Acta (BBA) 1694(1–3):81–95. https://doi.org/10.1016/j.bbamcr.2004.02.009
Voorhees RM, Fernández IS, Scheres SH, Hegde RS (2014) Structure of the mammalian ribosome-Sec61 complex to 3.4 Å resolution. Cell 157(7):1632–1643. https://doi.org/10.1016/j.cell.2014.05.024
Voorhees RM, Hegde RS (2016) Structure of the Sec61 channel opened by a signal sequence. Science (New York) 351(6268):88–91. https://doi.org/10.1126/science.aad4992
Wang S, Yang C-I, Shan S-O (2017) SecA mediates cotranslational targeting and translocation of an inner membrane protein. J Cell Biol. https://doi.org/10.1083/jcb.201704036
Wonderlin WF (2009) Constitutive, translation-independent opening of the protein-conducting channel in the endoplasmic reticulum. Eur J Physiol 457(4):917–930. https://doi.org/10.1007/s00424-008-0545-y
Woodbury RL, Hardy SJS, Randall LL (2002) Complex behavior in solution of homodimeric SecA. Protein Sci 11(4):875–882. https://doi.org/10.1110/ps.4090102
Yamane K, Ichihara S, Mizushima S (1987) In vitro translocation of protein across Escherichia coli membrane vesicles requires both the proton motive force and ATP. J Biol Chem 262(5):2358–2362
Zhang Q, Lahiri S, Banerjee T, Sun Z, Oliver D, Mukerji I (2017) Alignment of the protein substrate hairpin along the SecA two-helix finger primes protein transport in Escherichia coli. Proc Natl Acad Sci USA 114(35):S9343–S9348. https://doi.org/10.1073/pnas.1702201114
Zilberstein D, Schuldiner S, Padan E (1979) Proton electrochemical gradient in Escherichia coli cells and its relation to active transport of lactose. Biochemistry 18(4):669–673
Zimmer J, Nam Y, Rapoport TA (2008) Structure of a complex of the ATPase SecA and the protein-translocation channel. Nature 455(7215):936–943. https://doi.org/10.1038/nature07335
Acknowledgements
Open access funding provided by Austrian Science Fund (FWF). We thank Quentina Beatty for editorial help. We are indebted to the Austrian Science Fund for support (Grants #P 29841, #P 28213, #W1250 to DK and PP).
Funding
This study was funded by Austrian Science Fund, projects numbers #P 29841, #P 28213, #W1250 granted to DK and PP.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
Authors declare that they have no conflict of interest.
Ethical approval
This article does not contain any studies with human participants or animals performed by any of the authors.
Rights and permissions
Open Access This article is distributed under the terms of the Creative Commons Attribution 4.0 International License (http://creativecommons.org/licenses/by/4.0/), which permits unrestricted use, distribution, and reproduction in any medium, provided you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made.
About this article
Cite this article
Knyazev, D.G., Kuttner, R., Zimmermann, M. et al. Driving Forces of Translocation Through Bacterial Translocon SecYEG. J Membrane Biol 251, 329–343 (2018). https://doi.org/10.1007/s00232-017-0012-9
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00232-017-0012-9