Abstract
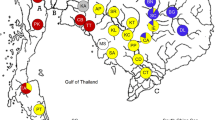
Brachidontes pharaonis (Fisher P, 1870) is an Indo-Pacific mussel that has colonized the Mediterranean Sea via the Suez Canal. Mussels may have migrated by natural dispersal of pelagic larvae, or they may have been transported on the hulls of ships, or in ballast water, or by some combination of these. Mitochondrial COI sequences (618 bp) from 101 mussels from six localities in the central and eastern Mediterranean Sea and from one site in the Red Sea were used to describe population structure. Analysis of molecular variance indicated that frequencies differed among populations, and that 92% of the variation resided within populations. The majority of haplotypes were private alleles. No simple pattern of longitudinal variation was detected for haplotype frequencies, haplotype diversity, or nucleotide diversity. A variety of tree-building algorithms (NJ, MP, ME) gave consistent results, showing two sister groups. One clade had a leucine (L-form) and the other one a methionine (M-form) at the 30th amino acid. These sympatric forms were detected within all localities and they had similar frequencies in males and females. The M- and L-form were separated by a 7.3% sequence divergence, indicating that this is an ancient polymorphism. Closely related species of mussels had exclusively the L-form, so we infer that the M-form evolved from the L-form lineage of B. pharaonis, probably in the northern portion of the Red Sea. Neutrality tests detected an excess of rare mutations, a pattern of variation expected in expanding populations. Patterns of variation detected by Tajima’s D suggest that some form of balancing selection has maintained this polymorphism.



Similar content being viewed by others
References
Apte S, Holland BS, Godwin LS, Gardner JPA (2000) Jumping ship: a stepping stone event mediating transfer of a nonindigenous species via a potentially unsuitable environment. Biol Invasions 2:75–79
Arcidiacono A, Di Geronimo I (1976) Studio biometrico di alcuni campioni di Brachidontes variabilis (Krauss). Conchiglie 12:61–74
Borsa P, Jarne P, Belkhir K, Bonhomme F (1994) Genetic structure of the palourde Ruditapes decussatus L., in the Mediterranean. In: Beaumont AR (ed) Genetics and evolution of aquatic organisms, Chapman and Hall, London, pp 103–113
Chemello R, Oliverio M (1995) Lessepsian migrations: a theoretical “island-jumping” model. Biol Mar Medit 3:444–446
Di Geronimo I (1971) Prima segnalazione nelle coste italiche di Brachidontes variabilis (Krauss). Boll Acc Gioenia Sci Nat Catania 10:847–852
Excoffier L, Smouse P, Quattro J (1992) Analysis of molecular variance inferred from metric distances among DNA haplotypes: application to human mitochondrial DNA restriction data. Genetics 131:479–491
Excoffier L, Smouse P (1994) Using allele frequencies and geographic subdivision to reconstruct gene genealogies within a species. Molecular variance parsimony. Genetics 136:343–359
Felsenstein J (1985) Confidence limits on phylogenies: an approach using the bootstrap. Evolution 39:783–791
Fisher P (1870) Sur la Faune conchyliologique marine des baies de Suez et de l’Akabah. Journal de Conchyliologie 13:178–179
Folmer O, Black M, Hoen W, Hoen W, Lutz R, Vrijenhoek R (1994) DNA primers for amplification of mitochondrial cytochrome c oxidase subunit I from diverse metozoan invertebrates. Mol Mar Biol Biotech 3:294–299
Fu YX (1997) Statistical tests of neutrality of mutations against population growth and background selection. Genetics 147:915–925
Galil BS (2000) A sea under siege–alien species in the Mediterranean. Biol Invasions 2:177–186
Garrigan D, Hedrick PW (2003) Delete period detecting adaptive molecular polymorphism: lessons from the MHC. Evolution 57:1707–1722
Gianguzza P, Chemello R, Riggio S (1997) Segnalazione di Brachidontes pharaonis (P. Fischer, 1870) (Bivalvia, Mytilidae) nella Salina di Marsala e considerazioni sulla distribuzione della specie in Mediterraneo. Boll Malacol 33:169–172
Hassan M, Harmelin-Vivien M, Bonhomme F (2003) Lessepsian invasion without bottleneck: example of two rabbitfish species (Siganus rivulatus and Siganus luridus). J Exp Mar Biol Ecol 291:219–232
Holland BS (2000) Genetics of marine bioinvasions. Hydrobiologia 420:63–71
Holland BS (2001) Invasion without a bottleneck: microsatellite variation in natural and invasive populations of the brown mussel Perna perna. Mar Biotech 3:407–415
Issel A (1869) Malacologia del Mar Rosso, Pisa pp 387
Kumar S, Tamura K, Nei M (2001) MEGA: molecular evolutionary genetic analysis, version 2.1. Pennsylvania State University University Park, Pennsylvania
Lee T, Ó Foighil D (2004) Hidden Floridian biodiversity: mitochondrial and nuclear gene trees reveal four cryptic species within the scorched mussel, Brachidontes exustus, species complex. Mol Ecol 13:3527–3542
Lee T, Ó Foighil D (2005) Placing the Floridian marine genetic disjunction into a regional evolutionary context using the scorched mussel, Brachidontes exustus, species complex. Evolution 59:2139–2158
Lewontin RC, Kojima K (1960) The evolutionary dynamics of complex polymorphisms. Evolution 14:458–472
McElroy D, Moran P, Bermingham E, Kornfield I (1992) REAP: an integrated environmental for the manipulation and phylogenetic analysis of restriction data. J Hered 83:157–158
Myers AA, Giller PS (1988) Process, pattern and scale in biogeography. In: Myers AA, Giller PS (eds) Analytical biogeography. Chapman & Hall, London, pp 3–12
Nei M (1987) Molecular evolutionary genetics. Columbia University Press, New York
Olson RR, Olson MH (1989) Food limitation of planktotrophic marine invertebrate larvae: does it control recruitment success? Ann Rev Ecol Syst 20:225–247
Por F D (1978) Lessepsian migration—the influx of Red Sea Biota into the Mediterranean by way of the Suez Canal. Ecological studies 23. Springer, Berlin Heidelberg New York
Rilov G, Gasith A, Benayahu Y (2002) Effect of an exotic prey on the feeding pattern of a predatory snail. Mar Env Res 54:85–98
Rawson PD, Burton RS (2002) Functional coadaptation between cytochrome c and cytochrome c oxidase within allopatric populations of a marine copepod. Proc Nat Acad Sci USA 99:12955–12958
Roff DA, Bentzen P (1989) The statistical analysis of mitochondrial DNA polymorphisms: chi-square (χ2) and the problem of small samples. Mol Biol Evol 6:539–545
Rozas J, Rozas R (1999) DnaSP version 3: an integrated program for molecular population genetics and molecular evolution analysis. Bioinformatics 15:174–175
Safriel UN, Gilboa A, Felsenburg T (1980) Distribution of rocky intertidal mussels in the Red Sea coasts of Sinai, the Suez Canal and the Mediterranean coast of Israel, with special reference to recent colonizers. J Biogeogr 7:39–62
Safriel UN, Ritte U (1983) Universal correlates of colonizing ability. In: Swingland IR, Greenwood PJ (eds) The ecology of animal movement. Clarendon, Oxford, pp 215–239
Saitou N, Nei M (1987) The neighbor-joining method: a new method for reconstructing phylogenetic trees. Mol Biol Evol 6:514–525
Sarà G, Romano C, Caruso M, Mazzola A (2000) The new Lessepsian entry Brachidontes pharaonis (Fisher P, 1870) (Bivalvia, Mytilidae) in the western Mediterranean: a physiological analysis under varying natural conditions. J Shell Res 19:967–977
Sarà G, Vizzini S, Mazzola A (2003) Sources of carbon and dietary habits of new Lessepsian entry Brachidontes pharaonis (Bivalvia, Mytilidae) in the western Mediterranean. Mar Biol 143:713–722
Schneider S, Excoffier L (1999) Estimation of demographic parameters from the distribution of pairwise differences when the mutation rates vary among sites: Application to human mitochondrial DNA. Genetics 152:1079–1089
Schneider S, Roessli D, Excoffier L (2000) Arlequin ver. 2.000: A software for Population Genetics Data Analysis. Genetics and Biometry Laboratory, University of Geneva, Switzerland
Shefer S, Abelson A, Mokady O, E Geffen (2004) Red to Mediterranean Sea bioinvasion: natural drift through the Suez Canal, or anthropogenic transport? Mol Ecol 13:2333–2343
Slatkin M (1985) Rare alleles as indicators of gene flow. Evolution 39:53–65
Stern S, Achituv Y (1977) Effects of temperature and salinity on the metabolism and byssal formation of Brachidontes variabilis Krauss (Bivalvia). Comp Bioch Phys 59A:101–105
Tajima F (1989) Statistical methods to test for nucleotide mutation hypothesis by DNA polymorphism. Genetics 123:585–595
Tamura K, Nei M (1993) Estimation of the number of nucleotide substitutions in the control region of mitochondrial DNA in humans and chimpanzees. Mol Biol Evol 10:512–526
Thompson JD, Higgins DG, Gibson TJ (1994) CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, positions-specific gap penalties and weight matrix choice. Nucleic Acids Res 22:4673–4680
Widdows J (1991) Physiological ecology of mussel larvae. Aquaculture 94:147–163
Wright S (1951) The genetical structure of populations. Ann Eugen 15:323–354
Acknowledgments
We thank Prof. Bella S Galil and Prof. Maoz Fine for collecting samples from Israeli coasts, and Prof. Pei-San Tsai for support in gonads examination. Research supported by University of Palermo (CORI and ex 60%).
Author information
Authors and Affiliations
Corresponding authors
Additional information
Communicated by R. Cattaneo-Vietti, Genova
Appendix
Appendix
Alignment of the variable sites among the 75 haplotypes of COI (618 bp) scored in B. pharaonis from the Mediterranean Sea and the Red Sea.
Sequences are listed dividing the two mitochondrial forms (L and M).


All haplotypes are compared with haplotype 1 M and are designated by a number (see also Table 1) and two letters: L indicating L-form or M, M-form, followed by m male, f female, n sex not identified.
N number of haplotypes scored in the Mediterranean samples (SA Salina di Marsala, T Termini Imerese, TN Torre Normanna, TA Tel Aviv, HA Haifa) and in the Red Sea (EG Safaga Bay)
aa1 and aa2, amino acid in haplotype 1 M and after mutation, respectively.
Rights and permissions
About this article
Cite this article
Sirna Terranova, M., Lo Brutto, S., Arculeo, M. et al. Population structure of Brachidontes pharaonis (P. Fisher, 1870) (Bivalvia, Mytilidae) in the Mediterranean Sea, and evolution of a novel mtDNA polymorphism. Mar Biol 150, 89–101 (2006). https://doi.org/10.1007/s00227-006-0330-4
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00227-006-0330-4