Abstract
The Ran GTPase protein is a guanine nucleotide-binding protein (GNBP) with an acknowledged profile in cancer onset, progression and metastases. The complex mechanism adopted by GNBPs in exchanging GDP for GTP is an intriguing process and crucial for Ran viability. The successful completion of the process is a fundamental aspect of propagating downstream signalling events. QM/MM molecular dynamics simulations were employed in this study to provide a deeper mechanistic understanding of the initiation of nucleotide exchange in Ran. Results indicate significant disruption of the metal-binding site upon interaction with RCC1 (the Ran guanine nucleotide exchange factor), overall culminating in the prominent shift of the divalent magnesium ion. The observed ion drifting is reasoned to occur as a consequence of the complex formation between Ran and RCC1 and is postulated to be a critical factor in the exchange process adopted by Ran. This is the first report to observe and detail such intricate dynamics for a protein in Ras superfamily.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
1 Introduction
Ran GTPase (Ran) is a small, 24 kDa eukaryotic protein belonging to the Ras superfamily of small G-proteins [1]. It exerts its function in many aspects of cellular activity [2]. Of the principal roles is the involvement in nucleocytoplasmic transport. Ran is also implicated in other cellular events including spindle assembly, coordination of DNA synthesis and nuclear envelope reformation. Ran is highly over-expressed in various cancer cells such as epithelial ovarian cancer and pancreatic cancer [3, 4]. It has been shown that silencing Ran decreased cell proliferation and cell adhesion in vitro [5]. Recently, El-Tanani et al. suggested that mutation of Ran is associated with activation of PI3K/Akt/mTORC1 and Ras/MEK/ERK pathways and therefore is a potential target for treatment of cancers [6].
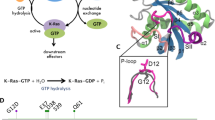
A fundamental aspect of finely tuned Ran regulation is the ‘molecular switch’ mechanism, i.e. the cycling of GTP (ON) and GDP (OFF) within the cell. In general, the GTPase transitions through three states, i.e. a GDP-bound state, an ‘empty’ state and a GTP-bound state [7, 8]. Maintenance and mediation of such a delicately equilibrated system are achieved through association of the GTPase with various accessory proteins. For the Ras superfamily, there are two key proteins employed in regulation of the GTP-bound/GDP-bound ratio. They are the guanine nucleotide exchange factor (GEF; RCC1 for Ran) and the GTPase activating protein (GAP; RanGAP for Ran). GEFs bind the GDP-bound form, increasing GDP dissociation, whereas GAPs bind the GTP-bound form and increase GTP hydrolysis [9]. Another class of effector proteins are the guanine nucleotide dissociation inhibitors (GDIs; NTF2 for Ran) [10]. These proteins bind the GDP-bound GTPase, essentially ‘locking’ the protein in an inactive, GDP-bound state. Their function in inhibiting GDP dissociation from the GTPase precludes nucleotide exchange, ultimately impeding GTP-mediated downstream signal transmission. For signal transduction with the ultimate aim of cell cycle progression, the GTPase must be turned ‘on’, which involves activation of the nucleotide exchange process. The initial step of nucleotide exchange is approach of the GEF. The GEF is then involved in disrupting the coordination of the magnesium system in the Ras superfamily, thereby culminating in breakdown of interactions between protein and the nucleotide system [11]. Unfortunately, the mechanism by which Ran executes its role in nucleotide exchange is not fully characterized or well understood. The most relevant crystal structure available for Ran nucleotide exchange is a complex of nucleotide-free Ran–RCC1 [12]. Resolution of this crystal structure provides a basis for nucleotide exchange by comparison with other available crystal structure information and coordinates. The crystal structure revealed RCC1 is comprised of a seven-bladed β-propeller and the Switch 2, α2 and α3 helices of Ran interact with RCC1 along the top of its β-propeller. A β-wedge protrusion from RCC1 was proposed to prevent the Switch 1 from adopting its GTP-bound conformation. This, although very useful, does not provide a dynamic vision of the transition between ON and OFF states. In attempting to understand the exchange process adopted by Ran, it is worth considering the previous studies regarding nucleotide exchange in other Ras superfamily members. Particularly, destabilization of magnesium coordination was strongly implied in the literature which was deemed as a determining factor in the exchange process [13–15]. Despite this, there appears to be significant diversity in the mechanistic action. For example, it was proposed that loss of the metal ion precedes major rearrangement of the Switch 1 region, which folds up and moves away from the metal- and base-binding site of an Mg2+-free form of GDP-bound RhoA. This jaw-like mechanism was suggested to be necessary for exchange, by increasing accessibility to the nucleotide. The interaction between Cdc42 and DOCK9 has also been discussed [16]. Authors suggested that opening of the Switch 1 loop aids exchange and also identified the α10 insert which orients into the nucleotide-binding site, utilizing a valine residue to block the metal-binding site which in turn stabilizes the nucleotide-free complex. Further nucleotide exchange activity in the ARA7–VPS9a was reported, suggesting that removal of the magnesium ion from the nucleotide-binding site of the GEF-bound GTPase is effected by a so-called ‘aspartate finger’ which inserts into the nucleotide-binding site, causing removal of the ion and formation of an interaction with the P-loop lysine [17]. This destabilizes the interaction of GDP with ARA7 GTPase. The nucleotide exchange in Arf1 as catalysed by the GEF, ARNO was reported, where computational modelling was employed to recreate intermediates in the nucleotide exchange pathway [18]. Previous reports imply the removal of magnesium as a prerequisite for the GEF interaction [19, 20]; however, the crystal structure of Arf1–GDP–BFA–GEF (e.g. PDB ID: 1s9d) implies that the Mg2+ ion is still present when the GEF binds [21]. This agrees with the model of GDP–GTPase–GEF containing the magnesium ion and subsequently promotes speculation about the specific mechanism involved in removal of the Mg2+ ion from the GEF-bound GTPase [22]. The relocation of the magnesium ion in RhoA was also discussed in another report where the repositioning of the ion between alpha and beta phosphates of GDP was suggested as a step prior to nucleotide dissociation [23]. Taking into account the previous literature, it is clear that the Ras superfamily displays significant diversity in the mechanistic action of GEFs. With Ran being a completely distinct family and comprising only a single member, it is important to consider all aspects of nucleotide exchange implied thus far, in addition to identifying possibly unique features of this protein–protein interaction. This study reports the first hybrid QM/MM molecular dynamics simulation targeted at identifying aspects relating to the nucleotide exchange of Ran.
2 Methods
2.1 Model generation
Two crystal structures were employed in the preparation of RanMgGDP–RCC1 complex model including the crystal structure of RanMgGDP (PDB ID: 3gj0) and the crystal structure of the regulator of chromosome condensation, RCC1 (PDB ID: 1a12) [24, 25]. The complex was modelled using the PDB entry 1i2m (a complex of nucleotide-free Ran–RCC1) as a template, which represents a complex post-GDP dissociation. Because the structure of RCC1 does not alter significantly upon complex formation with Ran [12], RCC1 was superimposed onto the RAN in order to generate the complex of RanMgGDP–RCC1. Firstly, RCC1 from PDB entry 1a12 was superimposed onto RCC1 from PDB entry 1i2m; Ran was then superimposed from the 3gj0 PDB entry onto the 1i2m complex. A complex of RanMgGDP bound with RCC1 was thus generated. The complex was minimized using the Desmond software [26], initially for 15 steps of SD minimization followed by a further 845 step minimization by the L-BFGS algorithm (convergence criteria set to 1.0 kcal/mol/Å).
The protein hydrogen atoms were removed in addition to all solvent molecules, except for the four water molecules coordinating the magnesium ion. Hydrogen atoms were added and appropriate 9520 water molecules and 4 chlorine counter ions were added to the RanMgGDP model system; the RanMgGDP–RCC1 system was solvated with 22,445 water molecules and neutralized with three sodium counter ions. Prior to MD the model is minimized using the PMEMD module in Amber with the ff99SB force field [27]. The system is first minimized with restraints placed on solute atoms. This minimization proceeds for 1000 steps of SD minimization and then switches to the CONJ algorithm for a further 4000 steps. In this second minimization, there are no restraints enforced and the model proceeds for 2500 steps of SD minimization, followed by a further 5000 steps of CONJ minimization. The resulting structure is used as the input for the subsequent MD simulations. A cut-off of 12 Å was employed for all non-bonded interactions in the minimizations.
2.2 Parameter preparation
Charges were calculated by optimizing a selected region by HF method. Optimization was first performed for RanMgGDP, for which the region included GDP, the magnesium ion and its complete first coordination sphere composing four waters, T24 and GDP. K23 was also included in the calculation as the large electrostatic contribution resides in close proximity to the Mg2+–GDP system. Residues T24 and K23 were truncated to keep the side chains and peptide backbones in the calculation. In total 91 atoms were included in the geometry optimization, which was performed with the Gaussian 03 program [28], using the restricted HF method with the 6–31 g (d) basis set.
Geometry optimization of the RanMgGDP–RCC1 follows the same procedure as that of the RanMgGDP system. For this model complex, a larger region was chosen for optimization (133 atoms), which included the magnesium ion, GDP, the four coordinating water molecules, the side chain of T24Ran, the side chain of K23Ran as well as three residues from the β-wedge of RCC1 (D148RCC1, N149RCC1 and N150RCC1). Amino acids were truncated similar to those in the Ran calculation.
Charge calculation was carried out in two stages: first electrostatic potential (ESP) charges for each atom were calculated and then these were converted to restrained electrostatic potential (RESP) charges [29]. RESP calculations were performed using the RESP program as part of the Amber suite of programs. The calculated charges for Mg–GDP in RanMgGDP and RanMgGDP–RCC1 systems are listed in supporting information Table S1 and S2, respectively. Denotation of the atoms is shown in supporting information Fig. S1.
2.3 MD simulations
The minimized system was heated to 300 K over a period of 50 ps. A weak restraint of 0.5 kcal/mol was placed on solute atoms. The simulation was executed with the PMEMD module of Amber, using periodic boundary conditions for 50,000 steps with a time step of 1 fs. The SHAKE algorithm was applied to all bonds involving hydrogen atoms and a cut-off of 12 Å was applied for non-bonded interactions.
After heating the system was subject to 1 ns conventional equilibration, where the system is allowed to stabilize at a pressure of 1 bar, over 1 ns. Using the PMEMD module, a time step of 1 fs was employed and periodic boundary conditions were applied. All restraints were removed, the SHAKE algorithm was applied to all bonds involving hydrogen, and a cut-off of 12 Å was used for the non-bonded interactions.
2.4 Hybrid QM/MM
Following equilibration, the system was subject to a further 1 ns equilibration using mixed QM/MM simulation techniques with the sander module of Amber. In the QM/MM calculation the magnesium ion, GDP, Thr24 and four coordinating water molecules were treated by PM3 method [30, 31]. The QM region was truncated to include the first coordination sphere of the divalent ion (Fig. S2). The total charge of the QM region is −1. The system was allowed to evolve and stabilize at a pressure of 1 bar for 1 ns with a time step of 2 fs. Both periodic boundary conditions and the SHAKE algorithm (for all bonds involving hydrogen) were applied. There was no restraint placed on the solute atoms. The boundary was treated via the link atom approach and a cut-off of 8 Å was used for non-bonded electrostatic interactions with an adapted implementation of the PME method.
After classical and hybrid QM/MM equilibration, the system was advanced to a QM/MM production simulation. The RanMgGDP complex was allowed to evolve over 15 ns under the QM/MM simulation protocol using the sander module. The system is set up with periodic boundary conditions and run for 15 ns, using a time step of 2 fs. The SHAKE algorithm is applied to all bonds involving hydrogen. There was no restraint placed on solute atoms and a cut-off of 8 Å was used for non-bonded electrostatic interactions.
Parameters used in RanMgGDP simulations are the same as those applied to simulations of the RanMgGDP–RCC1 model. The RanMgGDP–RCC1 system was allowed to evolve over 20 ns under the QM/MM simulation protocol, using a time step of 1 fs.
3 Results and discussion
3.1 Evolved stability of Ran from MD simulations
Because the N-terminal region of the Ran protein was flexible throughout the simulation, RMSD calculations were carried out for Ran, excluding the first seven residues.
Figure 1a shows the RMSD fluctuation in the alpha carbon atoms of the residues 8–207 in Ran throughout the 15 ns QM/MM production simulation. The simulation stabilizes at approximately 8 ns, remaining stable thereafter.
Figure 1b illustrates the fluctuation in alpha carbon RMSD throughout the RanMgGDP–RCC1 simulation. It can be seen that the complex system evolves to an RMSD of approximately 2 Å in the first 15 ns of simulation and then stabilizes, except that a sharp fluctuation in alpha carbon RMSD was observed towards the end of the production simulation. This fluctuation may correspond to conformational rearrangements of the complex system.
3.2 Conformational promiscuity
MD simulations with the RanMgGDP complex were executed to monitor the dynamic evolution of the species, as predicted in an aqueous environment. Starting from a well-defined crystal structure, we anticipate a fairly stable evolution during simulation, presupposing that the simulated protein is unlikely to undergo major rearrangements within the strict definition of simulation protocol. Indeed, this supposition holds true based on the RanMgGDP simulation, where results indicate a very stable system which refrains from dramatic rearrangement. Based on the premise of system stability, it is of considerable importance to highlight the stability of the usually ‘ill-defined’ magnesium and GDP throughout the QM/MM MD simulation. This justifies the parameters for magnesium and GDP and the starting model used for the MD simulations.
The secondary structure of RanMgGDP-RCC1 model is shown in Fig. S3. Switch 1 comprises residues 29–46 and Switch 2 comprises residues 67–76. The P-loop, composed of residues 17–25, was suggested to stabilize the nucleotide. The tip of the β-wedge comprises residues 147–150; among them, the two asparagines N149 and N150 are the most significant protruding residues. B-factors infer regions of flexibility and regions which are well ordered. The residue B-factors were calculated for the RanMgGDP simulated structure (Fig. S4). We observed the Switch 2 region exhibits considerable motion. In particular, the side chain of K71 is very flexible according to the B-factor calculation. This is reasonable considering the charged aliphatic lysine side chain and the polar solvent environment to which it is exposed. The Switch 2 region is known to be one of the most dynamic regions in Ras superfamily proteins and so increased B-factors in this region is not unexpected. Minor increases in B-factors are also observed for other regions in Ran; specifically, residues 34–38, 95–113, 127–143 and 179–207. Residues 34–38 comprise a section of the Switch 1 region in Ran, a region which is known to be extremely flexible. Residues 95–113 incorporate the helix α3 region in Ran. This region is exposed to the solvent environment and in this respect, it is reasonable to assume that there will be a level of flexibility in this region. Similarly, residues 127–143 reside close to the guanine nucleotide-binding site and incorporate helix α4 and so can react to conformational changes in and surrounding the GDP ligand.
The most notable aspect of the RanMgGDP–RCC1 simulation was the obvious relocation of the divalent magnesium ion. From the alpha carbon RMSD graph for RanMgGDP–RCC1 (Fig. 1b), considerable fluctuations were observed around 14–15 ns and between 19 and 21 ns. Up until 15 ns, the system is involved in initial system stabilization, where the RMSD of the alpha carbon atoms deviates to approximately 2 Å. After approximately 15 ns, the system settles into a relatively stable state, up until the point of magnesium shifting at approximately 18.5 ns. It is likely that the increased fluctuation is a result of the system adapting itself with the newly acquired location of magnesium and its coordinating partners. In this study, we focused on the residues surrounding the Mg2+-binding site as well as those surrounding the Switch 1 of Ran and the β-wedge in RCC1, because they are significant to Ran guanine nucleotide exchange.
The B-factors calculated for the core residues (i.e. res. 8–177) of Ran in the RanMgGDP–RCC1 protein (Fig. S5) imply the most significant motion appears to be in residues 25–50 and 120–140. These regions correspond to the Switch 1 (res. 29–46) as well as the base-binding site and the α4 helix region (res. 122–126 and res. 132–140, respectively). The Switch 1 is known to be flexible and the motion in the base-binding site is a result of the complex generation and motion surrounding the GDP ligand. The α4 helix resides on the RCC1-binding interface and the motion in this region is a result of complex formation. The calculated B-factors for RCC1 disclosed a region (res. 230–245) with increased flexibility compared to other residues (Fig. S6), which is a flexible, solvent-exposed RCC1 loop, located opposite to the Ran-interacting surface.
3.3 Conformational rearrangement: the fate of magnesium
Table 1 shows the distance evolution of magnesium and the β-phosphate oxygen of GDP between 18.500 and 18.550 ns. The distances for the first and last frames of the trajectory are also included as a reference. Significant disruption of the metal-coordinating conformations was observed in the simulations. The magnesium ion was observed to move away from the GDP ligand. Prior to the ion shift, the distance between magnesium and the β-phosphate of GDP remains very stable around 2.5 Å. The shift appears to occur rapidly and the Mg2+ ion continues to depart from the GDP β-phosphate oxygen atom after the initial ion–nucleotide bond cleavage. Therefore, we propose that the guanine nucleotide exchange in Ras superfamily proteins occurs through disruption of the interaction between magnesium and its ligating partners.
Figure 2 shows the motion of various elements throughout the simulation. Specifically, this depicts the evolution of GDP, E70Ran, E46Ran, the magnesium ion and the four coordinating water molecules over a period of 22.745 ns (Fig. 2). The bond cleavage between magnesium and GDP occurs after 18.525 ns (Fig. 2). To monitor this shift, the conformational changes in the residues surrounding this region are inspected throughout the simulation, specifically after 15 ns when the system appears moderately stabilized. A residue, E70Ran of Switch 2, shows considerable deviation when the magnesium ion is about to depart and seems to play significant role in the final interaction breakdown between the Mg2+–GDP system (Fig. 2). This residue is involved in a water-bridged hydrogen bond and salt bridge interactions and is thus important in retention of the metal-coordination system [32]. The carboxylate side chain of residue E70Ran orients into the metal-coordination site at the beginning of the simulation (Fig. 2). Thus, contacts are established between the metal ion and the coordinating waters, which is important for maintenance of the octahedral coordination observed in the Ran protein.
Major disruption of this metal-coordination site is observed after 18.500 ns and this coincides with a ‘flip’ of the glutamate side chain away from the metal-coordination site (Fig. 2). The side chain of E70Ran ‘flips’ between 18.515 and 18.525 ns preceding the shift of magnesium. This result is interesting in that it implies that the ‘flip’ is the determining factor in the interaction breakdown between magnesium and GDP. Despite the interaction with the metal-coordinated system being lost, the side chain of E70Ran does not appear to compensate this by further interactions with other solute molecules. Although it is clear that the magnesium system relocates during the simulation and that the side chain ‘flip’ of residue E70Ran is a major factor contributing to the final bond cleavage between magnesium and GDP, it is important to rationalize the post-shift location of this species. Visual analysis identified a residue, E46Ran, which proved to be important in permitting the successful transition of this magnesium-coordinated system (Fig. 2). The side chain of residue E46Ran is located far from the metal-coordination site at the beginning of the simulation. As the system evolves, the overhanging Switch 1 loop appears flexible, permitting reorientation of the E46Ran side chain to approach the magnesium centre occurring from 17.700 ns onwards (Fig. 2). As with the ‘flip’ of the side chain of E70Ran, this reorientation precedes the relocation of magnesium. Initially the terminal carboxylate group manoeuvres towards the P-loop region followed by gradual relaxation of the aliphatic chain towards the same region. The motion of this residue is suggested to be crucial for relocation of the magnesium system. Indeed, completion of a successful transition requires preparation of a suitable site in which residence can be assumed. Therefore, we propose that the conformational changes associated with this residue form the basis of a complementary metal-binding site. Upon relocation of the metal system, several significant contacts are established. The most notable is probably the salt bridge formed between the ion and carboxylate oxygen of E46Ran. In addition to this, E46Ran also forms water-bridged hydrogen bonds to the metal. The transition of E46Ran is stabilized by the concerted reorientation of K37Ran in the Switch 1 motif. Residue K37Ran maintains a strong salt bridge interaction with the carboxylate of E46Ran during the simulation.
Two residues seem be important for ensuring efficient metal relocation, namely T24 on the Ran P-loop and D65 on the Ran Switch 2. Residue T24Ran is included in the octahedrally composed first coordination sphere of the Mg2+ ion and close to the guanine nucleotide. Steadfast maintenance of the interaction between T24Ran and magnesium is evident throughout the simulation. It is proposed that retention of the interaction between magnesium and T24Ran throughout the simulation acts as a chaperone for the magnesium relocation. Residue D65Ran is also proposed to chaperone the metal system during relocation. The convenient location of this residue, at the beginning of the Switch 2 motif and close to the Switch 1 loop, is postulated to be critical for the effective relocation of the magnesium ion. The carboxylate side chain maintains salt bridge and water-bridged hydrogen bond interaction with magnesium which is in turn coordinated to the GDP. The salt bridge interaction is also sustained throughout the ion shift. This, together with T24Ran, can effectively function as an ‘escalator’ between the guanine nucleotide-binding site and the Switch 1 region, where contact with the magnesium coordination centre is maintained throughout the simulation.
It is also important to consider the influence of the β-wedge. N149RCC1 and N150RCC1 are in close proximity to the metal-coordination centre. The nucleotide and its surrounding Ran residues (e.g. K23Ran and E70Ran) provide significant electrostatic character to the metal-binding site. It is likely that the incoming β-wedge serves to dilute this strong electrostatic character. The neutralizing effect is provided by two asparagine residues N149 and N150 at the tip of the β-wedge (Fig. 2). These polar but neutral side chains orient close to the di-phosphate and function to detract a portion of the electronegativity manifested by GDP. The neutralizing effect is further enhanced by the terminating amino functionalities of the asparagine residues. Furthermore, we postulate that the relocation of E46Ran provides substantial electronegative character to enable comfortable relocation of the divalent ion, effectively compensating for the diminished electronegativity in the guanine nucleotide-binding site. Figure 2 illustrates the interference of the β-wedge residues in the Switch 2/P-loop interface in Ran. In addition to the electrostatic contribution, the β-wedge residues also provide a sterically challenging environment. Early accommodation of N149RCC1 between the P-loop and Switch 2 interface is apparent. Convenient insertion of the prominent β-wedge between the P-loop and Switch 2 regions increases the distance between these two motifs, driving them apart (Fig. 2). The distance between these two motifs increases such that tension on the magnesium system which stretches over these regions is created. D18Ran of the P-loop in Ran is identified to be crucial in the correct positioning of the β-wedge. From the simulation, we observed the formation of a strong salt bridge interaction between D18Ran and R147RCC1 on the β-wedge in RCC1, which may contribute significantly to the retention of the GEF. In view of the location and strength, it is possible that it acts as the linchpin for complex interaction, hinging both Ran and RCC1 interfaces. K71Ran, nested on the Switch 2 motif, is flexible and solvent exposed; interestingly, it was found to form a salt bridge with the carboxylate side chain of E200RCC1, which is close to the β-wedge. Although these interactions do not directly intrude on the metal site, they are likely important in maintaining the strong complex interaction between Ran and RCC1 proteins.
At this stage, it is worth summarizing the cooperative nature of the observed simulation results. In particular, the ion shift is indispensable in effecting the exchange process, and a concerted mechanism which permits the occurrence of this shift is outlined. Indeed, initiation of this process must first involve the approach of the GEF, RCC1, to the (correct) Ran interface. Convenient and direct insertion of β-wedge residues into the P-loop/Switch 2 interface of Ran is important for two reasons. Firstly, it is postulated that the residues in the P-loop and Switch 2 regions are crucial for defining effective complex formation. Binding of RCC1 to Ran in an irregular orientation likely renders the complex unproductive by the strict definition and evolution of protein architecture. Specifically, we propose that the strong salt bridge formed between D18Ran and R147RCC1 is critical in creation and maintenance of this protein–protein interaction. Secondly, the steric imposition originating from insertion of the pronounced β-wedge disrupts the finely tuned balance between the P-loop and Switch 2 components. In particular, N149RCC1 and N150RCC1 encroach on the metal-binding site and directly interact with the residues on the P-loop and Switch 2. This disrupts the electrostatic and steric balance experienced within the metal-binding site. Specific interaction of N149RCC1 with E70Ran strains the octahedrally coordinated magnesium system, whilst E46Ran reorients its side chain carboxylate above the P-loop/Switch 2 interface in order to prepare suitable environment to accommodate the metal ion. It appears that both a culmination of the increased hydrogen bond interaction between N149RCC1 and E70Ran and the increase in space and electronegative character introduced above the metal-binding site result in the ‘flip’ of E70Ran side chain, which could be viewed as ‘releasing the lock’ on magnesium coordination.
3.4 Implications for nucleotide exchange
Based on the available crystal structures in the literature [33] and our simulation results, the mechanism for initiation of RCC1-mediated nucleotide exchange in RanGTPase is proposed (Fig. 3) and the seamless perpetuation of this process within the cellular environment is hypothesized.
Overall, our simulation results imply that prominent disruption (of the P-loop/Switch 2 interface) by the β-wedge is not significant enough for instantaneous relocation of magnesium. We propose that attempts to equilibrate the electrostatic distribution and alleviate the steric challenge surrounding the metal–nucleotide site ultimately culminate in the ion shift, as the system reacts to the conformational changes upon complex formation (and evolution). In general, the initiation of nucleotide exchange in Ran can be analogized as a ‘disappearing act’ performed by the magnesium ion. The interaction of the β-wedge with Ran inevitably creates a sterically unfavourable environment, which strains the P-loop/Switch 2-balanced magnesium coordination system. It is likely that these impositions cause disruption to the finely tuned electrostatic character of the magnesium coordination system and the residues involved in the guanine nucleotide-binding site. This culminates in the orchestration of complete interaction breakdown between the Mg2+–GDP system.
It appears that a flip in residue E70Ran is involved in final breakdown of the interaction between Mg2+–GDP. Initially, the magnesium system is stabilized in the active site by residues of the P-loop (e.g. T24Ran) and Switch 2 (e.g. D65Ran and E70Ran). Insertion of β-wedge residues between the P-loop/Switch 2 interface perturbs the metal-binding site, encumbering the stable octahedral coordination. N149RCC1 appears critical for the disruption, as it forms a hydrogen bonding interaction with E70Ran, which removes a portion of the electronegativity initially provided by this residue. The interaction between the backbone of N150RCC1 and G20Ran also contributes to destabilization of the metal-binding site. This interaction, together with a culmination of the competition between N149RCC1and E70Ran and the steric effect imposed by insertion of N149RCC1 between the P-loop and Switch 2, ultimately result in collapse of the interaction between E70Ran and the Mg2+-coordinated system (by removal of the E70Ran side chain from the metal-binding site). Preparation of a suitable relocation site is achieved through reorientation of residue E46Ran to a position above the metal-binding site. The conformational shift is critical to provide an increased level of electronegativity into this region and the relocation is stabilized by a salt bridge interaction with K37Ran. In order to maintain a stable octahedrally coordinated metal system, the ion and coordination sphere relocate to the vicinity of Switch 1. The strong interaction maintained between the ion and residues T24Ran and D65Ran indicates that the metal system retains a level of stability but with threatened loss of its ‘comfortable’ octahedral coordination. Both T24Ran and D65Ran appear critical for the relocation, acting as ‘escalators’ or ‘hinges’ in chaperoning the metal system from the guanine nucleotide-binding site to the Switch 1 region in order to compensate for the loss of interaction between E70Ran and the metal-coordination system. The attractive carboxylate-rich environment and the abundance of space above the guanine nucleotide-binding site both appeal to the threatened octahedrally coordinated magnesium, and thus, the constraints within the GEF-bound P-loop/Switch 2 interface are alleviated to some extent. Subsequent to the ion shift, both D65Ran and E46Ran are within sufficient distance to form meaningful salt bridge interactions with the magnesium-coordinated system. The ion relocation causes loss of interaction between magnesium and GDP which is proposed to drastically reduce the affinity of Ran for GDP [34]. The reduction in affinity is suggested to be a major determinant in the interaction breakdown between the nucleotide and Ran. Given that release of magnesium decreases Ran affinity, it is possible that the phosphate terminus provides significant contribution to GDP-binding energy. It is suggested that loss of metal interaction causes the phosphate to ‘peel’ away from Ran, a process which may be aided by interaction with N150RCC1. Phosphate removal is likely accompanied by dislocation of the adjoining ribose and guanine motifs. The metal may completely dissociate from Ran after the shift, whereas it is also possible that the metal temporarily resides in the Switch 1 region during the nucleotide exchange process. In this situation, the relocation of the magnesium system not only alleviates spatial constraints in the P-loop/Switch 2 interface but also allows more space for the incoming GTP, post-GDP dissociation. With this, and the fact that the magnesium ion has strong association with the Switch 1 region post-shift, the events following GDP dissociation can be surmised. An incoming GTP molecule, as required for correct nucleotide exchange, is expected to bind into the guanine nucleotide-binding site and interact with the magnesium upon initial coordination. The location of the magnesium is useful for the incoming GTP molecule, being larger than the GDP molecule. Because of the interaction of the metal ion with the Switch 1 residues, it is postulated that magnesium moves to re-associate with the nucleotide into the catalytically important site upon GTP association. The interactions gained with Switch 1 through pre-exchange are strong enough to be maintained in the post-exchange ion relocation, which effectively pulls the Switch 1 region down with the metal, enveloping the guanine nucleotide-binding site (e.g. PDB ID: 2bku) [35]. The encapsulation by Switch 1 introduces steric interference with the interface residues of RCC1. Indeed, complete closure of the Switch 1 loop over GTP would clash with the RCC1 protein [12] and so it is hypothesized that this is the terminating factor in the nucleotide exchange process. Specifically, closure of Switch 1 down over the nucleotide-binding site signals the end of nucleotide exchange and interactions gained from this, between Ran and nucleotide, are sufficient to force the disengagement of the RCC1 protein. Indeed, the post-GDP dissociation hypothesis proposed here agrees with the concept of unsuccessful nucleotide exchange. In the nucleus, both GDP and GTP are present. It is therefore possible that upon initial GDP dissociation another GDP molecule could, in theory, associate. The lack of tri-phosphate is surmised to be insufficient in reaching the magnesium ion located by the Switch 1, and therefore, no valuable interaction would be established. In this case, the Switch 1 region would not ‘close’ and terminate nucleotide exchange; however, the GDP would simply dissociate and the process is perpetuated until a GTP associates and the exchange is successful.
A crystal structure of nucleotide- and magnesium-free Ran–RCC1 is available (PDB ID: 1i2m) which implies the absence of the GDP and metal prior to completion of exchange [12]. It is possible that, in addition to the dissociation of GDP, the interaction of magnesium and Ran may also be lost before an Mg2+–GTP system is invited into the binding site. It should be noted that the Switch 1 region in the crystal structure is not resolved. In particular, residues which are postulated to be involved in stabilizing the post-shift location of magnesium in this research are missing in the crystal structure. Therefore, we suggest Mg2+ is drifting during the nucleotide exchange, although the possibility of complete Mg2+ loss cannot be excluded.
In agreement with the results obtained in the present research the literature strongly implies destabilization of the magnesium coordination as a determining factor in the exchange process. Early literature concerning RhoA implies the loss of magnesium prior to the Switch 1 conformational change (i.e. closed to open) and GEF interaction [22, 36], but this cannot be the case for Ran. The Switch 1 region sandwiches the nucleotide in the GDP-bound form of RhoA (PDB ID: 1cc0); however, in the GDP-bound form of Ran the Switch 1 adopts the ‘open’ conformation (Fig. 4) [37]. Moreover, the Switch 1 ‘jaw’ in Ran is ‘open’ prior to GEF interaction (Fig. 4), as confirmed by the RanGDP–NTF2 complex (PDB ID: 1a2k) [38]. It is possible that RanGDP–NTF2 (PDB ID: 1a2k) is involved in preparing the template for the ion shifting. This hypothesis is drawn from various crystal structures (PDB IDs: 1a2k, 3gj0 and 1i2m). The structure of RanGDP–NTF2 represents a structure of Ran which is involved in the process of nucleocytoplasmic transport and precedes the exchange process. Furthermore, the NTF2 and RCC1 Ran-binding interfaces overlap, so NTF2 must dissociate prior to RCC1 interaction. The Switch 1 loop in all three complexes displays the ‘open’ conformation (Fig. 4), so it clear that the Switch 1 loop is ‘open’ prior to GEF interaction. The fact that the Switch 1 motif adopts the ‘open’ conformation prior to GEF interaction indicates that Ran employs a nucleotide exchange mechanism dissimilar to that of RhoA. Ran also appears to adopt a different nucleotide exchange mechanism than that represented in the Cdc42–DOCK9 and ARA7–VPS9a complexes [16, 39], which may be caused by the different GEF proteins. In addition, it is postulated that the Cdc42–DOCK9 requires a specific α10 motif which functions to block the magnesium-binding site and the ARA7–VPS9a interaction requests utilization of an ‘aspartate finger’ in forcing the removal of the ion. In our simulation results, the asparagine residues at the tip of the β-wedge insert close to the metal-coordination site; however, they do not appear to directly block the metal-binding site. Furthermore, an aspartate (or glutamate) residue responsible for the magnesium destabilization and/or loss was not observed.
Relocation of the metal ion concerning the RhoA–GEF mechanism has been suggested in recent literature [23]. Indeed, this corroborates with observations reported in this work, despite the direction of relocation being dissimilar. The difference between the ion drifting reported in the RhoA–GEF complex and our study can be rationalized by the difference in their Switch 1 conformations. In the RhoGDP structure, the Switch 1 region is folded down over the guanine nucleotide- and metal-binding site, i.e. the closed conformation (Fig. 4). As such, the region above the metal-binding site is significantly congested when compared to that of Ran, where the Switch 1 loop manifests an ‘open’ conformation in the GDP-bound state. It is surmised that the observed ion relocation between the alpha and beta phosphates of GDP in RhoA is due to the lack of available space above the metal-binding site, in addition to the apparently sufficient electronegativity provided by the alpha phosphate of GDP.
The concept of the magnesium drifting observed in the present work agrees with the rationale of the metal system disruption by corresponding GEFs. We presume that the incoming β-wedge in RCC1 has a neutralizing effect on the magnesium-binding site, where the disruption causes destabilization and subsequent relocation of the metal ion. In a similar respect, relocation towards the alpha phosphate (as in the case of RhoA), also indicates alteration of electrostatics in the original metal-binding site upon GEF interaction, resulting in the shift of the metal towards a more electronegative region [23]. We propose that the ‘disappearing act’ performed by the magnesium ion is critical for GDP dissociation and that the drastic conformational difference reported for the Ran Switch 1 loop is the driving force for the Switch 1-directed nucleotide exchange in Ran. Although it is theoretically possible that the Mg2+-free form of Ran constitutes another intermediate in the exchange process, in view of the conformation of the Switch 1 loop prior to GEF-binding and the convenient location of the ion drift, it is proposed that the ion initially resides in the vicinity of the Switch 1.
In order to understand the complete picture of nucleotide exchange mechanism of RanGTPase, we compared our simulation observations with the available crystal structure of Ran–RCC1 (PDB ID: 1i2m) to rationalize the transition between the model we obtained and the nucleotide-free Ran–RCC1 complex. In the complex, the Switch 1 loop is in an ‘open’ conformation with lack of defined coordinates. That is, the residues 32–37 in the Switch 1 loop (comprising residues 29–46Ran) were unresolved and missing in the crystal structure as well as residues 233–238 of RCC1. A common conception in the Ras-like GTPase nucleotide exchange mechanism is the stabilization of the nucleotide-free intermediate state by the interaction of the P-loop lysine residue (K23Ran) and either the aspartate at the beginning of Switch 2 or the glutamate at the end of the Switch 2 motif [19, 23, 39]. In the Ran–RCC1 crystal structure, K23Ran forms a salt bridge interaction with D65Ran on the Switch 2. Although in the MD study detailed in this research, the interaction of K23Ran with either D65Ran or E70Ran is not observed, it is possible that this interaction contributes to destabilization of GDP or it is gained upon GDP loss to stabilize the nucleotide-free state. Another difference in the reported guanine nucleotide exchange mechanism of Ran and other Ras-like GTPases is the lack of intrusion into the magnesium-binding site by residues from the GEF. In some GTPases, residues of the GEF have been reported to block the metal-binding site in the GDP-bound–GTPase–GEF complexes [14, 16, 39, 40]. However, this is not the case in the Ran–RCC1 complex interaction. A likely reason for this difference is the binding interfaces between the GTPase and its respective GEFs. Although the RCC1 interacts with the guanine nucleotide and the surroundings, the coverage is not significantly interfering with the metal-binding site. The RCC1 protein only interacts with the bottom face of Ran (PDB ID: 1i2m), whereas other GEFs cover a more extensive interface encompassing P-loop, Switch 1 and Switch 2 motifs (e.g. PDB ID’s: 1r8q, 3lw8, 1bkd and 1foe) [21, 23, 40, 41].
In the Ran–RCC1 crystal structure reported by Renault et al., K71Ran of the Switch 2 forms a salt bridge interaction with D18Ran of the P-loop [12]. In the models studied within our work this interaction is not observed. Firstly, in the MD simulated RanMgGDP structure, interaction formed between K71Ran and the solvent environment is observed. This is reasonable considering the solvent-exposed and polar nature of this residue. Upon generation of the RanMgGDP–RCC1 complex, this residue forms additional contacts with RCC1, including a salt bridge between K71Ran and E200RCC1. To rationalize the difference in the orientation of this residue between the RanMgGDP/RanMgGDP–RCC1 complexes and the Ran–RCC1 nucleotide-free complex, we suggest that the relocation of K71Ran occurs as a result of the dislodged magnesium ion. This would effectively remove a large portion of the electropositivity involved in stabilizing the guanine nucleotide within the binding site. Furthermore, in order to minimize GDP destabilization experienced by the Mg2+ ion drifting, the side chain of K71Ran flips in towards the metal-binding site. Since the nucleotide-free complex suggests a salt bridge interaction between K71Ran and D18Ran, it is likely the loss of GDP may permit formation of this interaction.
The crystal structure of the nucleotide-free Ran–RCC1 complex implies a large conformational shift in residues E70Ran and W104Ran [12]. Although the MD study carried out in the present research does not show the complete conformational change in these two residues, as defined in the literature [12], it is clear that residue E70Ran is removed from the metal-binding site (Fig. 5). Compared with the structure of the nucleotide-free Ran–RCC1 complex, we suggest the Switch 2 likely continues to depart from the metal-binding site. Further opening of the Switch 2/P-loop interface may cause a clash between the side chains of E70Ran and W104Ran, consequentially ‘flipping’ the large aromatic side chain. It will be interesting to uncover further aspects of this complex interaction, to further decipher characteristics of nucleotide exchange in Ran GTPase.
4 Conclusions
MD simulations have been employed in deciphering aspects of initiation of the nucleotide exchange process adopted by Ran GTPase. This is the first report where destabilization of the Mg2+–GDP system in Ras-like GTPases was observed through QM/MM MD techniques. These results provide a theoretical basis for the mechanism of initiation of nucleotide exchange in Ran. Further to providing structural information surrounding this intricate process, the novel methods employed in this research also provide a paradigm for future implementation of these modern techniques into investigating the biological process for such complex systems.
References
Melchior F (2001) Curr Biol 11:R257
Sazer S, Dasso M (2000) J Cell Sci 113:1111
Barrès V, Ouellet V, Lafontaine J, Tonin PN, Provencher DM, Mes-Masson AM (2010) Mol Cancer 9:272
Deng L, Lu Y, Zhao X, Sun Y, Shi Y, Fan H, Liu C, Zhou J, Nie Y, Wu K, Fan D, Guo X (2013) Biochem Biophys Res Commun 440:322
Kurisetty VV, Johnston PG, Johnston N, Erwin P, Crowe P, Fernig DG, Campbell FC, Anderson IP, Rudland PS, El-Tanani MK (2008) Oncogene 27:7139
Yuen HF, Chan KK, Grills C, Murray JT, Platt-Higgins A, Eldin OS, O’Byrne K, Janne P, Fennell DA, Johnston PG, Rudland PS, El-Tanani M (2012) Clin Cancer Res 18:380
Avis JM, Clarke PR (1996) J Cell Sci 109:2423
Lu S, Jang H, Muratcioglu S, Gursoy A, Keskin O, Nussinov R, Zhang J (2016) Chem Rev. doi:10.1021/acs.chemrev.5b00542
Cherfils J, Zeghouf M (2013) Physiol Rev 93:269
Yamada M, Tachibana T, Imamoto N, Yoneda Y (1998) Curr Biol 8:1339
Bos JL, Rehmann H, Wittinghofer A (2007) Cell 129:865
Renault L, Kuhlmann J, Henkel A, Wittinghofer A (2001) Cell 105:245
Sprang SR, Coleman DE (1998) Cell 95:155
Cherfils J, Chardin P (1999) Trends Biochem Sci 24:306
Pan JY, Wessling-Resnick M (1998) BioEssays 20:516
Yang J, Zhang Z, Roe SM, Marshall CJ, Barford D (2009) Science 325:1398
Uejima T, Ihara K, Goh T, Ito E, Sunada M, Ueda T, Nakano A, Wakatsuki S (2010) J Biol Chem 285:36689
Hamida-Rebaï ME, Robert CH (2010) PLoS One 5:e9142
Shimizu T, Ihara K, Maesaki R, Kuroda S, Kaibuchi K, Hakoshima T (2000) J Biol Chem 275:18311
Mori K, Hata M, Neya S, Hoshino T (2005) J Am Chem Soc 127:15127
Renault L, Guibert B, Cherfils J (2003) Nature 426:525
Zhang B, Zhang Y, Wang Z, Zheng Y (2000) J Biol Chem 275:25299
Klink BU, Barden S, Heidler TV, Borchers C, Ladwein M, Stradal TE, Rottner K, Heinz DW (2010) J Biol Chem 285:17197
Partridge JR, Schwartz TU (2009) J Mol Biol 391:375
Renault L, Nassar N, Vetter I, Becker J, Klebe C, Roth M, Wittinghofer A (1998) Nature 392:97
Maestro 9.0 ed, Schrödinger DES (2009) In: Schrodinger DES, Shaw DE (eds) Desmond research. Schrodinger, New York
Case DA, Darden TA, Cheatham TE III, Simmerling CL, Wang J, Duke RE, Luo R, Crowley M, Walker RC, Zhang W, Merz KM, Wang B, Hayik S, Roitberg A, Seabra G, Kolossvary I, Wong KF, Paesnai F, Vanicek J, Wu X, Brozell SR, Steinbrecher T, Gohlke H, Yang L, Tan C, Mongan J, Hornak V, Cui G, Mathews DH, Seetin MG, Sagui C, Babin V, Kollman PA (2008) AMBER10. University of California, San Francisco
Frisch MJ, Trucks GW, Schlegel HB, Scuseria GE, Robb MA, Cheeseman JR, Montgomery JA, Vreven T, Kudin KN, Burant JC, Millam JM, Iyengar SS, Tomasi J, Barone V, Mennucci B, Cossi M, Scalmani G, Rega N, Petersson GA, Nakatsuji H, Hada M, Ehara M, Toyota K, Fukuda R, Hasegawa J, Ishida M, Nakajima T, Honda Y, Kitao O, Nakai H, Klene M, Li X, Knox JE, Hratchian HP, Cross JB, Bakken V, Adamo C, Jaramillo J, Gomperts R, Stratmann RE, Yazyev O, Austin AJ, Cammi R, Pomelli C, Ochterski JW, Ayala PY, Morokuma K, Voth GA, Salvador P, Dannenberg JJ, Zakrzewski VG, Dapprich S, Daniels AD, Strain MC, Farkas O, Malick DK, Rabuck AD, Raghavachari K, Foresman JB, Ortiz JV, Cui Q, Baboul AG, Clifford S, Cioslowski J, Stefanov BB, Liu G, Liashenko A, Piskorz P, Komaromi I, Martin RL, Fox DJ, Keith T, Al-Laham MA, Peng CY, Nanayakkara A, Challacombe M, Gill PMW, Johnson B, Chen W, Wong MW, Gonzalez C, Pople JA (2004) Gaussian, Inc, Wallingford
Bayly CI, Cieplak P, Cornell WD, Kollman PA (1993) J Phys Chem 97:10269
Stewart JJP (1989) J Comput Chem 10:209
Stewart JJP (1991) J Comput Chem 12:320
Scheffzek K, Klebe C, Fritzwolf K, Kabsch W, Wittinghofer A (1995) Nature 374:378
Mott HR, Owen D (2015) Crit Rev Biochem Mol Biol 50:85
Klebe C, Nishimoto T, Wittinghofer F (1993) Biochemistry 32:11923
Lee SJ, Matsuura Y, Liu SM, Stewart M (2005) Nature 435:693
Rossman KL, Der CJ, Sondek J (2005) Nat Rev Mol Cell Biol 6:167
Longenecker K, Read P, Derewenda U, Dauter Z, Liu XP, Garrard S, Walker L, Somlyo AV, Nakamoto RK, Somlyo AP, Derewenda ZS (1999) Acta Crystallogr Sect D Biol Crystallogr 55:1503
Stewart M, Kent HM, McCoy AJ (1998) J Mol Biol 277:635
Uejima T, Ihara K, Goh T, Ito E, Sunada M, Ueda T, Nakano A, Wakatsuki S (2010) J Biol Chem 285:36689
Boriack-Sjodin PA, Margarit SM, Bar-Sagi D, Kuriyan J (1998) Nature 394:337
Worthylake DK, Rossman KL, Sondek J (2000) Nature 408:682
Acknowledgments
The authors are grateful for the computing resources from QUB high-performance computing centre. MCM acknowledges the Department of Employment and Learning and the Centre for Cancer Research and Cell Biology for funding the research. We are also grateful to Dr Mohamed El-Tanani and Dr Marie Migaud from the Centre for Cancer Research and Cell Biology at Queen’s University Belfast for the helpful discussion.
Author information
Authors and Affiliations
Corresponding author
Additional information
Published as part of the special collection of articles “Charge Transfer Modeling in Chemistry”.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
Open Access This article is distributed under the terms of the Creative Commons Attribution 4.0 International License (http://creativecommons.org/licenses/by/4.0/), which permits unrestricted use, distribution, and reproduction in any medium, provided you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made.
About this article
Cite this article
Mooney, M.C., Xu, Y., McClory, J. et al. A disappearing act performed by magnesium: the nucleotide exchange mechanism of Ran GTPase by quantum mechanics/molecular mechanics studies. Theor Chem Acc 135, 197 (2016). https://doi.org/10.1007/s00214-016-1953-2
Received:
Accepted:
Published:
DOI: https://doi.org/10.1007/s00214-016-1953-2