Abstract
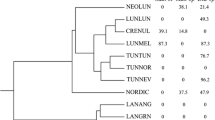
Spartina alterniflora is the dominant flowering plant of regularly flooded salt marshes along the Atlantic and Gulf coasts of the United States. Studies have suggested that there may be a genetic basis for the morphological, physiological, and phenological differences observed in plants sampled over a broad geographic range. Randomly amplified polymorphic DNA (RAPD) analysis was employed to assess the genetic variability in tall formS. alterniflora from the Atlantic and Gulf coasts. Twenty-nine RAPD primers produced 300 scoreable electrophoretic bands, of which 225 were polymorphic (75%). An UPGMA (unweighted pair-group method using an arithmetic average) phenogram based on Jaccard’s genetic distances showed three clusters of plants: New England/New Jersey, North Carolina/South Atlantic, and Gulf coast, Analysis of molecular variance (AMOVA) was used to estimate how genetic variability is partitioned among regions, areas, and individuals. The resulting variance components were highly significant at all hierachical levels for the sampling regime employed. The correlation between genetic and estimated coastal geographic flow among plants from geographically separate areas is not probable due to differences in flowering phenology, a total barrier to the exchange of genetic information is not likely. Present data and results of previous studies suggest a genetic continuum for this species rather than discrete, isolated populations.
Similar content being viewed by others
Literature Cited
Anderson, C. M. and M. Treshow. 1980. A review of environmental and genetic factors that affect height inSpartina alterniflora Loisel. (salt marsh cord grass). Estuaries 3:168–176.
Doyle, J. J. and J. L. Doyle. 1987. A rapid DNA isolation procedure for small quantities of fresh leaf tissue. Phytochemical Bulletin 19:11–15.
Excoffier, L., P. E. Smouse, and J. M. Quatro. 1992. Analysis of molecular variance inferred from metric distances among DNA haplotypes: application to human mitochondrial DNA restriction data. Genetics 131:479–491.
Felsenstein, J. 1993. PHYLIP (Phylogeny Inference Package) version 3.5c. Distributed by the author Department of Geneties, University of Washington. Seattle, WA, USA.
Freshwater, D. W. 1988. Relative genome-size differences among populations ofSpartina alterniflora Loisel. (Poaccac) along east and gulf coasts of U.S.A.. Journal of Experimental Marine Biology and Ecology 120:239–246.
Freshwater, D. W. and J. Rueness. 1994. Phylogenetic relationships of some EuropeanGelidium (Gelidiales. Rhodophyta) species, based onrhel. nucleotide sequence analysis. Phycologia 33:187–194.
Gallagher, J. L., G. F. Somers, D. M. Grant, and D. M. Seliskar. 1988. Persistent differences in two forms ofSpartina alterniflora: a common garden experiment. Ecology 69:1005–1008.
Huff, D. R., R. Peakall, and P. E. Smouse. 1993. RAPD variation within and among natural populations of outcrossing buffalograss [Buchloë dactyloides (Nutt.) Engelm.]. Theoretical and Applied Genetics 86:927–934.
Jaccard, P. 1908. Nouvelles recherches sur la distribution florale. Bulletin. Societe Vaudorse des Science Naturelles 44:223–270.
Kapraun, D. F. 1997. GENESTAT-3, A computer program for calculating Jaccard’s and Euclidean distances. Kapraun Computational Solution, Apex, NC, USA.
Maddison, W. P. and D. R. Maddison. 1992. MacClade: Analysis of phylogeny and character evolution. Version 3.01. Sinauer Associates. Sunderland, MA, USA.
Mantel N. A. 1967. The detection of disease clustering and a gencralized regression approach. Cancer Research 27:209–220.
Page, R. D. M. 1996. Tree View: An application to display phylogenetic trees on personal computers. Computer Applications in the Biosciences 12:357–358
Raymond M. and F. Rousset. 1995. GENEPOP (version 1.2): population geneties software for exact tests and ecumenicisim. Journal of Heredity 86:248–249.
Seneca, E. D. 1974. Germination and seedling response of Atlantic and Gulf coasts populations ofSpartina alterniflora. American Journal of Botany 61:947–956.
Silander, J. A. 1979. Microevolution and clone structure inSpartina patens. Science 203:658–660.
Silander, J. A. 1984. The genetic basis of the ecological amplitude ofSpartina patens. III. Allozyme variation. Botanical Gazette 145:569–577.
Silander, J. A. 1985. The genetic basis of the ecological amplitude ofSpartina patens. II. Variance and correlation analysis. Evolution 39:1034–1052.
Slatkin, M. 1993. Isolation by distance in equilibrium and non-equilibrium populations. Evolution 47:264–279.
Somers, G. F. and D. Grant. 1981. Influence of seed source upon phenology of flowering ofSpartina alterniflora Loisel and the likelihood of cross pollination. American Journal of Botany 68: 6–9.
Stiller, J. W. and A. L. Denton. 1995. One hundred years ofSpartina alterniflora (Poaceae) in Willapa Bay, Washington random amplified polymorphic DNA analysis of an invasive population. Molecular Ecology 4:355–363.
van Oppen, M. J., H. Klerk, M. de Graaf, W. Stam and J. L. Olsen. 1996. Assessing the limits of random amplified polymorphie DNAs (RAPDs) in seaweed biogeography. Journal of Phycology 32:433–444.
Vivian-Smith, G. and E. W. Stiles. 1994. Dispersal of salt marsh seeds on the feet and feathers of waterfowl. Wetlands 14:316–319.
Webb, S. D. 1990. Historical biogeography. p. 84–85.In R. L. Myers and J. J. Ewel (eds.) Ecosystems of Florida. The University of Central Florida Press, Orlando, FL, USA.
Welsh, J. and M. McClelland. 1990. Fingerprinting genomes using PCR with arbitrary primers. Nucleic Acids Research 24:7213–7218.
Author information
Authors and Affiliations
Rights and permissions
About this article
Cite this article
O’Brien, D.L., Freshwater, D.W. Genetic diversity within tall formSpartina alterniflora Loisel along the Atlantic and Gulf coasts of the United States. Wetlands 19, 352–358 (1999). https://doi.org/10.1007/BF03161766
Received:
Revised:
Accepted:
Issue Date:
DOI: https://doi.org/10.1007/BF03161766