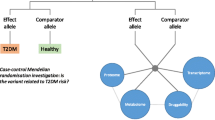
Abstract
Diabetes and its complications cause a heavy disease burden globally. Identifying exposures, risk factors and molecular processes causally associated with the development of diabetes can provide important evidence bases for disease prevention and spur novel therapeutic strategies. Mendelian randomisation (MR), an epidemiological approach that uses genetic instruments to infer causal associations between an exposure and an outcome, can be leveraged to complement evidence from observational and clinical studies. This narrative review aims to summarise the evidence on potential causal risk factors for diabetes by integrating published MR studies on type 1 and 2 diabetes, and to reflect on future perspectives of MR studies on diabetes. Despite the genetic influence on type 1 diabetes, few MR studies have been conducted to identify causal exposures or molecular processes leading to increased disease risk. In type 2 diabetes, MR analyses support causal associations of somatic, mental and lifestyle factors with development of the disease. These studies have also identified biomarkers, some of them derived from the gut microbiota, and molecular processes leading to increased disease risk. These studies provide valuable data to better understand disease pathophysiology and explore potential therapeutic targets. Because genetic association studies have mostly been restricted to participants of European descent, multi-ancestry cohorts are needed to examine the role of different types of physical activity, dietary components, metabolites, protein biomarkers and gut microbiome in diabetes development.
Graphical abstract

Similar content being viewed by others


Introduction
Diabetes is a leading health issue that causes severe disease and has a huge economic burden worldwide [1, 2]. Many epidemiological studies have assessed the causes of diabetes to provide an evidence base for disease prevention. For example, in type 2 diabetes, an exposure-wide umbrella review including 142 factors identified a wide range of biomarkers, medical conditions and dietary, lifestyle, environmental and psychosocial factors that were associated with the risk of disease [3]. The picture is somewhat different for type 1 diabetes owing to the strong genetic contribution and less influence of external factors. In addition to genetic factors, only a few environmental factors, including birthweight and childhood obesity, have been linked to type 1 diabetes [4]. While results from observational studies have provided initial evidence of potential exposures associated with diabetes, residual confounding and reverse causation limit our understanding of the complex set of factors underlying the development of diabetes. Thus, whether the factors observed in previous observational studies are causally associated with the risk of diabetes remains unconfirmed. A clear appraisal of the causal risk factors for diabetes is of great importance for disease prevention.
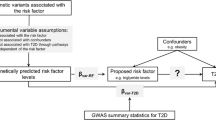
Mendelian randomisation (MR) is an epidemiological method that can strengthen causal inference by using genetic variants as instrumental variables [5]. An instrumental variable is a variable that satisfies three main conditions: (1) it is associated with the exposure (relevance assumption); (2) it does not share a common cause with the outcome (independence assumption); and (3) it is related to the outcome only through the exposure (exclusion restriction assumption) (Fig. 1). The text box summarises the common terms used in MR studies and their key concepts and limitations. As genetic variants are randomly assorted at conception and thus are generally unassociated with environmental and self-adopted factors, MR is believed to be less affected by measured and unmeasured confounding factors. This narrative review aims to summarise the evidence on potential causal risk factors for diabetes by integrating published MR studies on type 1 and 2 diabetes, and to reflect on future perspectives of MR studies on diabetes.
Study design and assumptions of MR analysis. The process of MR analysis is shown from top to bottom. In detail, MR analysis is based on genome-wide association analyses of the exposure and outcome. Genetic instruments for the exposure are independent SNPs that are strongly associated with the exposure of interest in a genome-wide association analysis in an unselected sample, such as a general population. Likewise, summary-level data on the outcome are obtained from a genome-wide association analysis of a binary phenotype that defines the population into cases and controls. The directed acyclic graph represents the study design and assumptions of MR analysis; G indicates the genetic instruments, X indicates the exposure of interest, Y indicates the outcome of interest, and U indicates the confounders. There are three important assumptions in MR analysis. Assumption 1 indicates that the genetic variants used as the instrumental variable should be robustly associated with the exposure. Assumption 2 indicates that the instrumental variable should not be associated with any confounders. Assumption 3 indicates that the instrumental variable used should affect the risk of the outcome only through the risk factor, not through alternative pathways. Regarding causal inference, the MR design resembles that of an RCT; specifically, the random allocation of genetic variants in MR mimics the randomisation process of RCTs, which minimises confounding effects. Source: Manhattan plot reproduced from Ikram et al [75], available under a CC BY 2.5 licence (https://creativecommons.org/licenses/by/2.5/). This figure is available as a downloadable slide
Causal exposures and risk factors for type 1 diabetes
Because there is a strong genetic component in type 1 diabetes, MR studies of type 1 diabetes are limited and only a few potentially modifiable risk factors have been identified (Table 1). Low birthweight [6], childhood obesity [6, 7] and a higher abundance of the Bifidobacterium genus [8] have been associated with an increased risk of type 1 diabetes. MR studies have found no associations of adult body size [6], features of the liver or pancreas [9] and serum 25-hydroxyvitamin D levels [10] with type 1 diabetes.
A protein-wide MR study examined the associations of 1611 circulating protein biomarkers with the risk of type 1 diabetes and identified associations for signal regulatory protein gamma, IL-27 Epstein–Barr virus-induced 3 and chymotrypsinogen B1 [11]. These findings linking certain viral infections, particularly by enteroviruses (e.g. coxsackievirus), with the risk of type 1 diabetes are consistent with recent observational studies [12], thus providing an avenue to better understand and prevent this disease.
Causal exposures and risk factors for type 2 diabetes
Most MR studies on glycaemic outcomes have focused on type 2 diabetes. Our previous exposure-wide MR study examined the associations of 97 exposures with risk of type 2 diabetes using data from the DIAbetes Genetics Replication And Meta-analysis (DIAGRAM) consortium (74,124 cases and 824,006 controls). In total, 34 factors that were possibly causally associated with the risk of type 2 diabetes were identified [13]. In Table 2 we summarise and update the associations of a wide range of exposures with type 2 diabetes from MR studies on diabetes.
Somatic and psychological health status
The results of MR studies of somatic and psychological health status in relation to type 2 diabetes are summarised in Table 2. Contradictory associations were reported for LDL-cholesterol and type 2 diabetes, with an inverse association observed in a European population and a positive association in an African population [13,14,15]. A recent study further identified that the diabetogenic effect of low levels of LDL-cholesterol might be mediated by increased BMI [16]. Lower levels of bilirubin (a marker of liver function) [17], testosterone [18] and thyrotropin [19] were associated with an increased risk of type 2 diabetes in some MR studies, but not all [13, 20,21,22]. Sex-specific associations were observed for testosterone [23, 24], with an increased risk of type 2 diabetes in women but a decreased risk in men with higher testosterone levels [23]. Insomnia, but no other sleep-related traits, was associated with type 2 diabetes [13].
Adiposity-related factors
Similar to the large body of evidence from prospective observational studies, childhood obesity, adulthood overall obesity and central obesity, excessive liver fat and whole-body and visceral fat mass were all associated with an increased risk of type 2 diabetes [9, 25,26,27,28,29]. Plasma levels of adiponectin, an adipocyte-secreted hormone, are decreased in individuals with obesity, which was associated with an increased risk of type 2 diabetes [30]. However, this association was inconsistent in MR sensitivity analyses [30], suggesting that the association may be biased by pleiotropy (e.g. from fat mass). Several MR studies have found that lower birthweight, independent of adult body weight, is associated with a higher risk of type 2 diabetes [31], which may suggest a role of the uterine environment and fetal development in the development of type 2 diabetes.
Lifestyle and nutritional factors
MR studies have strengthened the causal role of cigarette smoking in type 2 diabetes and failed to convincingly confirm the effects of physical activity and alcohol and coffee consumption on type 2 diabetes risk [13, 32, 33]. Although alcohol consumption instrumented by 83 SNPs was not associated with type 2 diabetes, the main SNP that associates with higher alcohol consumption and alcohol abuse in European populations (i.e. rs1229984 in the ADH1B gene) was significantly associated with an increased risk of disease [13]. A robust inverse association between coffee consumption and type 2 diabetes risk has been reported in many observational studies [34]. However, genetically predicted higher coffee consumption was not associated with a decreased risk of type 2 diabetes in MR studies [13, 35]. Pleiotropic effects of the SNPs used may cause this lack of association (e.g. from fat mass or other hot beverages or caffeine-containing drinks) and the inverse relationship between genetically proxied coffee consumption and plasma caffeine levels (i.e. the genetic variants with the strongest association with higher coffee consumption are associated with lower plasma caffeine levels) [36].
An MR study found an inverse association between circulating 25-hydroxyvitamin D levels and type 2 diabetes risk [37], and this association might be driven by the vitamin D synthesis pathway [37,38,39]. Lower levels of vitamin K1 (phylloquinone) [40] and higher levels of iron [41] were associated with an increased risk of type 2 diabetes. Eight out of ten plasma fatty acids were found to be associated with type 2 diabetes; however, the associations, with the exception of palmitoleic acid, were driven by SNPs in the FADS1/2 genes [42]. Thus, whether these associations were biased by this pleiotropic gene, which encodes a key enzyme in fatty acid metabolism, remains unknown [43].
Despite the popularity of MR studies for investigating dietary and lifestyle exposures in diabetes and cardiometabolic diseases, there are unique challenges in such studies of these time-varying, compositional and intercorrelated exposures [44]. For example, MR analyses of nutritional exposures based on genetic instruments for a single measure of diet collected in midlife bear an underlying assumption that, on average, the dietary assessment tool is representative of long-term habitual intake. Furthermore, like many behavioural exposures, nutrition is intercorrelated with numerous other lifestyle and environmental factors. Recent studies have documented that confounding and reverse causation affecting traditional epidemiological studies may also impact genetic associations [45]. A recent study has shown that half of the genetic variants associated with diet are the consequence of increased BMI and that it is possible to use genetics to correct for confounding and reverse causation to strengthen genetic correlations and causal inference [45].
IGF-1 and inflammatory biomarkers
Genetically predicted elevated levels of IGF-1, a peptide hormone similar in molecular structure to insulin, were positively associated with the risk of type 2 diabetes [46]. Given the heterogeneous effects of IGF-1-associated SNPs on type 2 diabetes, a recent MR analysis examined several clusters of IGF-1-associated SNPs in relation to type 2 diabetes and specified that this overall positive association might be explained by pathways related to amino acid metabolism and genomic integrity [47]. However, the main cluster of IGF-1-associated SNPs that were associated with a decreased risk of type 2 diabetes mapped to the growth hormone signalling pathway [47], possibly mediated by pleiotropic effects from fat mass, as growth hormone secretion is decreased in obesity [48].
As for inflammatory biomarkers, the IL-1 and IL-6 pathways may be involved in the development of type 2 diabetes [13, 49], even though the evidence is weak. One additional minor allele of the IL6R SNP rs7529229 (corresponding to the effect of taking tocilizumab 4–8 mg/kg every 4 weeks) was suggestively associated with a reduced risk of type 2 diabetes (OR 0.97, 95% CI 0.94, 1.00), which implied a possible role of IL-6 receptor blockade in type 2 diabetes prevention.
Circulating metabolites and proteins
One of the first demonstrations of the use of MR to study circulating metabolites was in relation to the previously reported epidemiological association between plasma levels of branched-chain amino acids (BCAAs) and the risk of type 2 diabetes [50]. In an MR analysis using genetic variation at the PPM1K locus (which encodes a mitochondrial phosphatase that activates branched-chain α-ketoacid dehydrogenase [BCKD]), an increase in leucine, isoleucine and valine levels was associated with an increased odds of type 2 diabetes [50]. However, given that BCKD has a range of substrates besides leucine, isoleucine and valine, untangling which of these substrates causes type 2 diabetes is challenging. A separate MR analysis of BCAAs showed that higher BCAA levels have no causal effects on insulin resistance but, rather, genetically raised insulin resistance drives higher circulating fasting BCAA levels [51]. A metabolome-wide MR approach confirmed evidence of the strong reverse causal effect, indicating that the genetic predisposition to type 2 diabetes may trigger early changes in valine and leucine [52]. Other products of amino acid catabolism, such as 2-aminoadipic acid (2-AAA) or α-hydroxybutyrate, are strongly associated with incident type 2 diabetes in observational studies [53], but MR studies have failed to demonstrate evidence of causality [54]. There are many reasons for the discrepancies between observational studies and MR studies, but the fact that observational studies have been conducted in a mixture of individuals with normoglycaemia and impaired glucose tolerance could explain these differences. A study in the Framingham cohort restricted to individuals with strict normoglycaemia at baseline (fasting glucose <5.6 mmol/l) provided evidence of a subset of 19 metabolites associated with the risk of diabetes among apparently healthy individuals [55]. Pathway enrichment analyses and MR showed that metabolites in the nitrogen metabolism pathway are causally related to the development of diabetes [55].
Integration of genomic and small molecule data across platforms enables the discovery of regulators of human metabolism and translation into clinical insights. A recent genome-wide meta-analysis of 174 metabolite levels across six cohorts, including up to 86,507 participants, identified ~500 genetic loci influencing metabolite levels [56]. Among many relevant findings for dysglycaemia, the study provided evidence that a missense p.Asp470Asn (rs17681684) variant in the GLP2R gene, which encodes the receptor for glucagon-like peptide 2, was associated with a 4% higher type 2 diabetes risk. Findings from a metabolome-wide MR analysis further identified new metabolites that potentially play a causal role in type 2 diabetes, including betaine, glutamic acid, lysine, alanine and mannose [52].
High-throughput detection and quantification of serum proteins in a large human population can provide insight into the molecular processes underlying diabetes risk. A protein-wide MR study examined the associations of 164 proteins with genome-wide association summary statistics available from the independent INTERVAL study and identified 16 proteins as potentially having a causal effect on the development of type 2 diabetes [57]. A recent protein-wide MR study examined the associations of 1089 circulating protein biomarkers with the risk of type 2 diabetes [58]. The analyses identified 20 proteins that might be causally associated with type 2 diabetes. These findings may provide evidence to support therapeutic development in type 2 diabetes.
MR studies on circulating metabolites and proteins usually employ a cis-variant located in an encoding gene region as the instrumental variable, which satisfies three key assumptions of MR. However, these MR associations can still be influenced by the genome-wide associations analyses on metabolites and proteins as well as corresponding profiling process (possible bias caused by batch effects) [59] and different high-throughput platforms [60]. Of note, using cis-variants as instrumental variables may not always completely rule out horizontal pleiotropy, especially when one gene regulates several metabolites and proteins that are not in a common pathway. In this case, multivariable MR analysis or removing the pleiotropic SNPs may help reduce this bias.
Gut microbiota and related metabolites
With increasing evidence suggesting that the human gut microbiome plays a role in immune function and metabolic disease, there is a need to discriminate between microbiome features that are causal for disease and those that are a consequence of disease or its treatment. A study including genome-wide genetic data, gut metagenomic sequencing and measurements of faecal short-chain fatty acids showed that a host genetic-driven increase in gut production of butyrate was associated with improved insulin response following an oral glucose test. In contrast, abnormalities in the production or absorption of propionate were causally related to an increased risk of type 2 diabetes [61]. Another two-sample MR study identified seven genera of gut microbiota nominally associated with type 2 diabetes [62]. For gut microbiota-related metabolites, a separate study found that genetically predicted higher trimethylamine N-oxide and carnitine levels were not associated with higher odds of type 2 diabetes. However, the study found possible associations of high choline and low betaine levels with an increased risk of type 2 diabetes [63]. Of note, although many genome-wide association analyses of the gut microbiome have been carried out, high-quality MR studies on the gut microbiome in relation to diabetes are limited [8]. This may raise doubt over the applicability of host genetic variants as an instrumental variable to mimic the function of the gut microbiome.
Assessment of included MR studies on diabetes
The overall quality of the MR studies included was satisfactory, with careful genetic instrument selection criteria, comparatively large sample sizes and different approaches to testing the robustness of the findings. As for the examination of the assumptions of MR, assumption 1 was usually found to be satisfied by using genetic variants associated with the exposure of interest at the genome-wide significance level. However, there was no unified threshold for linkage disequilibrium of SNPs. Using a high or low threshold of linkage disequilibrium could lead to an inflated rate of type 1 and 2 errors, respectively. As MR analysis can minimise confounding, the associations are less likely to be biased by confounding but cannot be completely immune to this bias, especially when genetic instruments have large pleiotropy effects. Except for studies using individual-level data, whether genetic instruments were primarily associated with other phenotypes or were associated with confounders was rarely examined in these MR studies. The most common bias in MR analysis is horizontal pleiotropy caused by violation of assumption 3, the exclusion restriction assumption, which means that genetic variants affect the outcome through alternative pathways, not only through the exposure of interest. The associations with type 1 and 2 diabetes summarised in this review were robust in sensitivity analyses, and most studies used MR-Egger or MR pleiotropy residual sum and outlier (MR-PRESSO) to detect potential horizontal pleiotropy. Of note, even though statistical methods can detect and minimise the influence of horizontal pleiotropy, instrumental variable selection is a crucial process for reducing the bias. Using genetic variants in genes with well-understood biological functions as instrumental variables usually satisfies the assumptions of MR analysis and thus generates precise and correct associations. However, it is difficult to identify specific genetic variants for certain exposures, especially for health behaviours and complex phenotypes. Therefore, a thoughtful examination of pleiotropy should be conducted in analyses using multiple genetic instruments. Evidence from observational studies and clinical trials should be used in interpreting MR findings. Robust MR findings, in turn, should be examined in clinical trials. In addition, it is tricky to interpret MR results, especially for binary exposures. Given that the exposure in MR analysis is not an exact phenotype but is proxied by the effects of genetic variants on a certain trait, this genetically proxied exposure usually mimics a lifetime chronic effect, which hinders the exploration of time-specific associations.
Future perspectives
-
The null findings in previous MR studies may have been caused by inadequate power, particularly for weak associations of exposures proxied by a few SNPs that explained a small phenotypic variance. For exposures with robust associations in traditional observational studies, the neutral associations in MR studies deserve to be re-examined in well-powered studies with robust genetic instruments for the exposures and large sample sizes for the diabetes outcomes.
-
Most previous MR studies were based on summary-level data, which do not allow the exploration of potential non-linear associations (e.g. J- or U-shaped); rather, it can only be assumed that the association is linear without a threshold effect. MR analysis using individual-level data from large-scale biobanks and studies is needed to examine the non-linearity of the associations.
-
More effort should be put into MR studies on non-heritable exposures or exposures without genetic association information. For example, MR analyses of the association of diet and physical activity with diabetes risk are warranted.
-
Most MR studies have been based on data from European populations. With more and more data available from other populations, such as Asian and African populations, future MR studies are encouraged to include data from multi-ancestry cohorts.
-
Even though the associations between protein biomarkers and diabetes risk were examined in a few MR studies [11, 58], more independent verification is needed to confirm these findings. In addition, the intermediate roles of blood proteins and metabolites in the pathways from environmental exposure to diabetes should be investigated to provide evidence for treatment and intervention.
-
Even though many statistical approaches, such as the weighted median, MR-Egger, MR-PRESSO, MR-Cluster and contamination mixture methods, have been developed to detect pleiotropy and verify the association with different assumptions, more efforts are needed to generate new statistical approaches to handle pleiotropy and other limitations.
Conclusion
This review has integrated data from published MR studies on type 1 and 2 diabetes to highlight the many possible causal risk factors for dysglycaemia. While few studies have been conducted for type 1 diabetes, most MR analyses support that social, demographic, metabolic and lifestyle factors are causally associated with the development of type 2 diabetes. More MR studies in multi-ancestry cohorts are needed to examine the role of diet in the development of diabetes. MR investigations based on data on metabolites, protein biomarkers and the gut microbiome may help to illustrate the pathological molecular basis of diabetes.
Abbreviations
- BCAA:
-
Branched-chain amino acid
- BCKD:
-
Branched-chain α-ketoacid dehydrogenase
- MR:
-
Mendelian randomisation
References
Ferguson D, Finck BN (2021) Emerging therapeutic approaches for the treatment of NAFLD and type 2 diabetes mellitus. Nat Rev Endocrinol 17(8):484–495. https://doi.org/10.1038/s41574-021-00507-z
Sun H, Saeedi P, Karuranga S et al (2022) IDF diabetes atlas: global, regional and country-level diabetes prevalence estimates for 2021 and projections for 2045. Diabetes Res Clin Pract 183:109119. https://doi.org/10.1016/j.diabres.2021.109119
Bellou V, Belbasis L, Tzoulaki I, Evangelou E (2018) Risk factors for type 2 diabetes mellitus: an exposure-wide umbrella review of meta-analyses. PLoS One 13(3):e0194127. https://doi.org/10.1371/journal.pone.0194127
Rewers M, Ludvigsson J (2016) Environmental risk factors for type 1 diabetes. Lancet 387(10035):2340–2348. https://doi.org/10.1016/s0140-6736(16)30507-4
Burgess S, Thompson SG (2015) Mendelian randomization: methods for using genetic variants in causal estimation. Chapman and Hall/CRC, London, UK
Richardson TG, Crouch DJM, Power GM et al (2022) Childhood body size directly increases type 1 diabetes risk based on a lifecourse Mendelian randomization approach. Nat Commun 13(1):2337. https://doi.org/10.1038/s41467-022-29932-y
Censin JC, Nowak C, Cooper N, Bergsten P, Todd JA, Fall T (2017) Childhood adiposity and risk of type 1 diabetes: a Mendelian randomization study. PLoS Med 14(8):e1002362. https://doi.org/10.1371/journal.pmed.1002362
Xu Q, Ni JJ, Han BX et al (2021) Causal relationship between gut microbiota and autoimmune diseases: a two-sample Mendelian randomization study. Front Immunol 12:746998. https://doi.org/10.3389/fimmu.2021.746998
Martin S, Sorokin EP, Thomas EL et al (2022) Estimating the effect of liver and pancreas volume and fat content on risk of diabetes: a Mendelian randomization study. Diabetes Care 45(2):460–468. https://doi.org/10.2337/dc21-1262
Manousaki D, Harroud A, Mitchell RE et al (2021) Vitamin D levels and risk of type 1 diabetes: a Mendelian randomization study. PLoS Med 18(2):e1003536. https://doi.org/10.1371/journal.pmed.1003536
Yazdanpanah N, Yazdanpanah M, Wang Y et al (2022) Clinically relevant circulating protein biomarkers for type 1 diabetes: evidence from a two-sample Mendelian randomization study. Diabetes Care 45(1):169–177. https://doi.org/10.2337/dc21-1049
Op de Beeck A, Eizirik DL (2016) Viral infections in type 1 diabetes mellitus--why the β cells? Nat Rev Endocrinol 12(5):263–273. https://doi.org/10.1038/nrendo.2016.30
Yuan S, Larsson SC (2020) An atlas on risk factors for type 2 diabetes: a wide-angled Mendelian randomisation study. Diabetologia 63(11):2359–2371. https://doi.org/10.1007/s00125-020-05253-x
Fall T, Xie W, Poon W et al (2015) Using genetic variants to assess the relationship between circulating lipids and type 2 diabetes. Diabetes 64(7):2676–2684. https://doi.org/10.2337/db14-1710
Soremekun O, Karhunen V, He Y et al (2022) Lipid traits and type 2 diabetes risk in African ancestry individuals: a Mendelian randomization study. EBioMedicine 78:103953. https://doi.org/10.1016/j.ebiom.2022.103953
Wu P, Moon JY, Daghlas I et al (2022) Obesity partially mediates the diabetogenic effect of lowering LDL cholesterol. Diabetes Care 45(1):232–240. https://doi.org/10.2337/dc21-1284
Abbasi A, Deetman PE, Corpeleijn E et al (2015) Bilirubin as a potential causal factor in type 2 diabetes risk: a Mendelian randomization study. Diabetes 64(4):1459–1469. https://doi.org/10.2337/db14-0228
Syed AAS, He L, Shi Y (2020) The potential effect of aberrant testosterone levels on common diseases: a Mendelian randomization study. Genes (Basel) 11(7):721. https://doi.org/10.3390/genes11070721
Kuś A, Marouli E, Del Greco MF et al (2021) Variation in normal range thyroid function affects serum cholesterol levels, blood pressure, and type 2 diabetes risk: a Mendelian randomization study. Thyroid 31(5):721–731. https://doi.org/10.1089/thy.2020.0393
Mohammadi-Shemirani P, Chong M, Pigeyre M, Morton RW, Gerstein HC, Paré G (2020) Effects of lifelong testosterone exposure on health and disease using Mendelian randomization. Elife 9:e58914. https://doi.org/10.7554/eLife.58914
Hou L, Li H, Si S et al (2021) Exploring the causal pathway from bilirubin to CVD and diabetes in the UK biobank cohort study: observational findings and Mendelian randomization studies. Atherosclerosis 320:112–121. https://doi.org/10.1016/j.atherosclerosis.2020.12.005
Bos MM, Smit RAJ, Trompet S, van Heemst D, Noordam R (2017) Thyroid signaling, insulin resistance, and 2 diabetes mellitus: a Mendelian randomization study. J Clin Endocrinol Metab 102(6):1960–1970. https://doi.org/10.1210/jc.2016-2816
Ruth KS, Day FR, Tyrrell J et al (2020) Using human genetics to understand the disease impacts of testosterone in men and women. Nat Med 26(2):252–258. https://doi.org/10.1038/s41591-020-0751-5
Yuan S, Wang L, Sun J et al (2022) Genetically predicted sex hormone levels and health outcomes: phenome-wide Mendelian randomization investigation. Int J Epidemiol. https://doi.org/10.1093/ije/dyac036
Geng T, Smith CE, Li C, Huang T (2018) Childhood BMI and adult type 2 diabetes, coronary artery diseases, chronic kidney disease, and cardiometabolic traits: a Mendelian randomization analysis. Diabetes Care 41(5):1089–1096. https://doi.org/10.2337/dc17-2141
Wainberg M, Mahajan A, Kundaje A et al (2019) Homogeneity in the association of body mass index with type 2 diabetes across the UK Biobank: a Mendelian randomization study. PLoS Med 16(12):e1002982. https://doi.org/10.1371/journal.pmed.1002982
Dale CE, Fatemifar G, Palmer TM et al (2017) Causal associations of adiposity and body fat distribution with coronary heart disease, stroke subtypes, and type 2 diabetes mellitus: a Mendelian randomization analysis. Circulation 135(24):2373–2388. https://doi.org/10.1161/circulationaha.116.026560
Karlsson T, Rask-Andersen M, Pan G et al (2019) Contribution of genetics to visceral adiposity and its relation to cardiovascular and metabolic disease. Nat Med 25(9):1390–1395. https://doi.org/10.1038/s41591-019-0563-7
Larsson SC, Burgess S (2020) Fat mass and fat-free mass in relation to cardiometabolic diseases: a two-sample Mendelian randomization study. J Intern Med 288(2):260–262. https://doi.org/10.1111/joim.13078
Nielsen MB, Çolak Y, Benn M, Nordestgaard BG (2021) Low plasma adiponectin in risk of type 2 diabetes: observational analysis and one- and two-sample Mendelian randomization analyses in 756,219 individuals. Diabetes 70(11):2694–2705. https://doi.org/10.2337/db21-0131
Huang T, Wang T, Zheng Y et al (2019) Association of birth weight with type 2 diabetes and glycemic traits: a Mendelian randomization study. JAMA Netw Open 2(9):e1910915. https://doi.org/10.1001/jamanetworkopen.2019.10915
Meisinger C, Linseisen J, Leitzmann M, Baurecht H, Baumeister SE (2020) Association of physical activity and sedentary behavior with type 2 diabetes and glycemic traits: a two-sample Mendelian randomization study. BMJ Open Diabetes Res Care 8(2):e001896. https://doi.org/10.1136/bmjdrc-2020-001896
Yuan S, Larsson SC (2019) A causal relationship between cigarette smoking and type 2 diabetes mellitus: a Mendelian randomization study. Sci Rep 9(1):19342. https://doi.org/10.1038/s41598-019-56014-9
Carlström M, Larsson SC (2018) Coffee consumption and reduced risk of developing type 2 diabetes: a systematic review with meta-analysis. Nutr Rev 76(6):395–417. https://doi.org/10.1093/nutrit/nuy014
Wang X, Jia J, Huang T (2022) Coffee types and type 2 diabetes mellitus: large-scale cross-phenotype association study and Mendelian randomization analysis. Front Endocrinol (Lausanne) 13:818831. https://doi.org/10.3389/fendo.2022.818831
Cornelis MC, Munafo MR (2018) Mendelian randomization studies of coffee and caffeine consumption. Nutrients 10(10):1343. https://doi.org/10.3390/nu10101343
Yuan S, Jiang X, Michaelsson K, Larsson SC (2019) Genetic prediction of serum 25-hydroxyvitamin D, calcium, and parathyroid hormone levels in relation to development of type 2 diabetes: a Mendelian randomization study. Diabetes Care 42(12):2197–2203. https://doi.org/10.2337/dc19-1247
Lu L, Bennett DA, Millwood IY et al (2018) Association of vitamin D with risk of type 2 diabetes: a Mendelian randomisation study in European and Chinese adults. PLoS Med 15(5):e1002566. https://doi.org/10.1371/journal.pmed.1002566
Afzal S, Brøndum-Jacobsen P, Bojesen SE, Nordestgaard BG (2014) Vitamin D concentration, obesity, and risk of diabetes: a Mendelian randomisation study. Lancet Diabetes Endocrinol 2(4):298–306. https://doi.org/10.1016/s2213-8587(13)70200-6
Zwakenberg SR, Remmelzwaal S, Beulens JWJ et al (2019) Circulating phylloquinone concentrations and risk of type 2 diabetes: a Mendelian randomization study. Diabetes 68(1):220–225. https://doi.org/10.2337/db18-0543
Wang X, Fang X, Zheng W et al (2021) Genetic support of a causal relationship between iron status and type 2 diabetes: a Mendelian randomization study. J Clin Endocrinol Metab 106(11):e4641–e4651. https://doi.org/10.1210/clinem/dgab454
Yuan S, Larsson SC (2020) Association of genetic variants related to plasma fatty acids with type 2 diabetes mellitus and glycaemic traits: a Mendelian randomisation study. Diabetologia 63(1):116–123. https://doi.org/10.1007/s00125-019-05019-0
Jäger S, Cuadrat R, Hoffmann P, Wittenbecher C, Schulze MB (2020) Desaturase activity and the risk of type 2 diabetes and coronary artery disease: a Mendelian randomization study. Nutrients 12(8):2261. https://doi.org/10.3390/nu12082261
Merino J, Tobias DK (2022) The unique challenges of studying the genetics of diet and nutrition. Nat Med 28(2):221–222. https://doi.org/10.1038/s41591-021-01626-w
Pirastu N, McDonnell C, Grzeszkowiak EJ et al (2022) Using genetic variation to disentangle the complex relationship between food intake and health outcomes. PLoS Genet 18(6):e1010162. https://doi.org/10.1371/journal.pgen.1010162
Larsson SC, Michaëlsson K, Burgess S (2020) IGF-1 and cardiometabolic diseases: a Mendelian randomisation study. Diabetologia 63(9):1775–1782. https://doi.org/10.1007/s00125-020-05190-9
Wang W, Tesfay EB, van Klinken JB et al (2022) Clustered Mendelian randomization analyses identify distinct and opposing pathways in the association between genetically influenced insulin-like growth factor-1 and type 2 diabetes mellitus. Int J Epidemiol. https://doi.org/10.1093/ije/dyac119
Frystyk J, Brick DJ, Gerweck AV, Utz AL, Miller KK (2009) Bioactive insulin-like growth factor-I in obesity. J Clin Endocrinol Metab 94(8):3093–3097. https://doi.org/10.1210/jc.2009-0614
Swerdlow DI, Holmes MV, Kuchenbaecker KB et al (2012) The interleukin-6 receptor as a target for prevention of coronary heart disease: a mendelian randomisation analysis. Lancet 379(9822):1214–1224. https://doi.org/10.1016/s0140-6736(12)60110-x
Lotta LA, Scott RA, Sharp SJ et al (2016) Genetic predisposition to an impaired metabolism of the branched-chain amino acids and risk of type 2 diabetes: a Mendelian randomisation analysis. PLoS Med 13(11):e1002179. https://doi.org/10.1371/journal.pmed.1002179
Mahendran Y, Jonsson A, Have CT et al (2017) Genetic evidence of a causal effect of insulin resistance on branched-chain amino acid levels. Diabetologia 60(5):873–878. https://doi.org/10.1007/s00125-017-4222-6
Porcu E, Gilardi F, Darrous L et al (2021) Triangulating evidence from longitudinal and Mendelian randomization studies of metabolomic biomarkers for type 2 diabetes. Sci Rep 11(1):6197. https://doi.org/10.1038/s41598-021-85684-7
Wang TJ, Ngo D, Psychogios N et al (2013) 2-Aminoadipic acid is a biomarker for diabetes risk. J Clin Invest 123(10):4309–4317. https://doi.org/10.1172/jci64801
Shi M, Wang C, Mei H et al (2022) Genetic architecture of plasma alpha-aminoadipic acid reveals a relationship with high-density lipoprotein cholesterol. J Am Heart Assoc 11(11):e024388. https://doi.org/10.1161/jaha.121.024388
Merino J, Leong A, Liu CT et al (2018) Metabolomics insights into early type 2 diabetes pathogenesis and detection in individuals with normal fasting glucose. Diabetologia 61(6):1315–1324. https://doi.org/10.1007/s00125-018-4599-x
Lotta LA, Pietzner M, Stewart ID et al (2021) A cross-platform approach identifies genetic regulators of human metabolism and health. Nat Genet 53(1):54–64. https://doi.org/10.1038/s41588-020-00751-5
Gudmundsdottir V, Zaghlool SB, Emilsson V et al (2020) Circulating protein signatures and causal candidates for type 2 diabetes. Diabetes 69(8):1843–1853. https://doi.org/10.2337/db19-1070
Ghanbari F, Yazdanpanah N, Yazdanpanah M, Richards JB, Manousaki D (2022) Connecting genomics and proteomics to identify protein biomarkers for adult and youth-onset type 2 diabetes: a two-sample Mendelian randomization study. Diabetes 71(6):1324–1337. https://doi.org/10.2337/db21-1046
Leek JT, Scharpf RB, Bravo HC et al (2010) Tackling the widespread and critical impact of batch effects in high-throughput data. Nat Rev Genet 11(10):733–739. https://doi.org/10.1038/nrg2825
Pietzner M, Wheeler E, Carrasco-Zanini J et al (2021) Synergistic insights into human health from aptamer- and antibody-based proteomic profiling. Nat Commun 12(1):6822. https://doi.org/10.1038/s41467-021-27164-0
Sanna S, van Zuydam NR, Mahajan A et al (2019) Causal relationships among the gut microbiome, short-chain fatty acids and metabolic diseases. Nat Genet 51(4):600–605. https://doi.org/10.1038/s41588-019-0350-x
Yang Q, Lin SL, Kwok MK, Leung GM, Schooling CM (2018) The roles of 27 genera of human gut microbiota in ischemic heart disease, type 2 diabetes mellitus, and their risk factors: a Mendelian randomization study. Am J Epidemiol 187(9):1916–1922. https://doi.org/10.1093/aje/kwy096
Jia J, Dou P, Gao M et al (2019) Assessment of causal direction between gut microbiota-dependent metabolites and cardiometabolic health: a bidirectional Mendelian randomization analysis. Diabetes 68(9):1747–1755. https://doi.org/10.2337/db19-0153
Haycock PC, Burgess S, Nounu A et al (2017) Association between telomere length and risk of cancer and non-neoplastic diseases: a Mendelian randomization study. JAMA Oncol 3(5):636–651. https://doi.org/10.1001/jamaoncol.2016.5945
Aikens RC, Zhao W, Saleheen D et al (2017) Systolic blood pressure and risk of type 2 diabetes: a Mendelian randomization study. Diabetes 66(2):543–550. https://doi.org/10.2337/db16-0868
Sluijs I, Holmes MV, van der Schouw YT et al (2015) A Mendelian randomization study of circulating uric acid and type 2 diabetes. Diabetes 64(8):3028–3036. https://doi.org/10.2337/db14-0742
Zhang K, Jia Y, Wang R et al (2022) Rheumatoid arthritis and the risk of major cardiometabolic diseases: a Mendelian randomization study. Scand J Rheumatol 1–7. https://doi.org/10.1080/03009742.2022.2070988
De Silva NMG, Borges MC, Hingorani AD et al (2019) Liver function and risk of type 2 diabetes: bidirectional Mendelian randomization study. Diabetes 68(8):1681–1691. https://doi.org/10.2337/db18-1048
Zhu T, Cui J, Goodarzi MO (2021) Polycystic ovary syndrome and risk of type 2 diabetes, coronary heart disease, and stroke. Diabetes 70(2):627–637. https://doi.org/10.2337/db20-0800
Wang F, Wang J, Li Y et al (2019) Gallstone disease and type 2 diabetes risk: a Mendelian Randomization study. Hepatology 70(2):610–620. https://doi.org/10.1002/hep.30403
Cao M, Cui B (2020) Negative effects of age at menarche on risk of cardiometabolic diseases in adulthood: a Mendelian randomization study. J Clin Endocrinol Metab 105(2):dgz071. https://doi.org/10.1210/clinem/dgz071
Tang B, Yuan S, Xiong Y, He Q, Larsson SC (2020) Major depressive disorder and cardiometabolic diseases: a bidirectional Mendelian randomisation study. Diabetologia 63(7):1305–1311. https://doi.org/10.1007/s00125-020-05131-6
Vissers LET, Sluijs I, van der Schouw YT et al (2019) Dairy product intake and risk of type 2 diabetes in EPIC-InterAct: a Mendelian randomization study. Diabetes Care 42(4):568–575. https://doi.org/10.2337/dc18-2034
Noordam R, Oudt CH, Bos MM, Smit RAJ, van Heemst D (2018) High-sensitivity C-reactive protein, low-grade systemic inflammation and type 2 diabetes mellitus: a two-sample Mendelian randomization study. Nutr Metab Cardiovasc Dis 28(8):795–802. https://doi.org/10.1016/j.numecd.2018.03.008
Ikram MK, Xueling S, Jensen RA et al (2010) Four novel loci (19q13, 6q24, 12q24, and 5q14) influence the microcirculation in vivo. PLoS Genet 6(10):e1001184. https://doi.org/10.1371/journal.pgen.1001184
Authors’ relationships and activities
JM is an Associate Editor for Diabetologia but played no role in evaluating this manuscript.
Contribution statement
All authors were responsible for drafting the article and revising it critically for important intellectual content. All authors approved the version to be published.
Funding
Open access funding provided by Karolinska Institute.
Author information
Authors and Affiliations
Corresponding author
Additional information
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Figure slide
(PPTX 151 kb)
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Yuan, S., Merino, J. & Larsson, S.C. Causal factors underlying diabetes risk informed by Mendelian randomisation analysis: evidence, opportunities and challenges. Diabetologia 66, 800–812 (2023). https://doi.org/10.1007/s00125-023-05879-7
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00125-023-05879-7