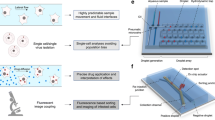
Abstract
Viral infectious diseases can erupt rapidly and are capable of causing massive health issues. Recent evidence on their high specificity over host species and cell type have fueled research into decoding host–viral interaction. The conventional approach relies on cell culturing-plate and multiwell plate-based analysis, which not only utilizes a large amount of reagents but are also population biased and gives ensemble measurement of biological assay. The animal model does not perfectly recapitulate human disease nor do they provide a point of care analysis. Studying the direct interaction between the virus and the host cell remain challenging. The researcher, therefore, has addressed these challenges using a microfluidic device, which promises high-throughput analysis, analysis at the single-cell level, and small consumption of reagents. These features intensify the understanding of various biological interactions in terms of single-cell heterogeneity in a rapid, accurate, and cost-effective manner. While conventional methods will continue to serve biology and virology well, we envisage that microfluidic platforms for viral infections at a single-cell level will be an impressive method to develop an antiviral drug, discover essential proteins for infection, and to understand viral mutants. This chapter emphasizes the various micro and nanodevice developed for the detection and capture of viruses. In addition, it also deals with an understanding of host–viral interactions and comments on how this understanding might give profound resolution to biological research as well as to the biomedical approach.
Similar content being viewed by others
References
Alivisatos P (2004) The use of nanocrystals in biological detection. Nat Biotechnol 22(1):47–52
Au SH et al (2013) Cellular bias on the microscale: probing the effects of digital microfluidic actuation on mammalian cell health, fitness and phenotype. Integr Biol 5(8):1014–1025
Bae JE, Kim IS (2010) Multiplex PCR for rapid detection of minute virus of mice, bovine parvovirus, and bovine herpesvirus during the manufacture of cell culture-derived biopharmaceuticals. Biotechnol Bioprocess Eng 15:1031–1037
Ben-Ari Ya et al (2013) Microfluidic large scale integration of viral–host interaction analysis. Lab Chip 13(12):2202–2209
Cao YC, Jin R, Mirkin CA (2002) Nanoparticles with Raman spectroscopic fingerprints for DNA and RNA detection. Science 297(5586):1536–1540
Chang Y-F et al (2010) Detection of swine-origin influenza A (H1N1) viruses using a localized surface plasmon coupled fluorescence fiber-optic biosensor. Biosens Bioelectron 26(3):1068–1073
Chen DL, Gerdts CJ, Ismagilov RF (2005) Using microfluidics to observe the effect of mixing on nucleation of protein crystals. J Am Chem Soc 127(27):9672–9673
Choi K et al (2012) Digital microfluidics. 5(1):413–440
Cui C, Griffiths A, Li G, Silva LM, Kramer MF, Gaasterland T, Wang XJ, Coen DM (2006) Prediction and identification of herpes simplex virus 1-encoded microRNAs. J Virol 80(11):5499–5508
DaPalma T et al (2010) A systematic approach to virus–virus interactions. Virus Res 149(1):1–9
Diaz-Munoz SL (2017) Viral coinfection is shaped by host ecology and virus–virus interactions across diverse microbial taxa and environments. Virus Evol 3(1)
Dougan JA et al (2007) Enhanced oligonucleotide–nanoparticle conjugate stability using thioctic acid modified oligonucleotides. Nucleic Acids Res 35(11):3668–3675
Draz MS et al (2012) Hybrid nanocluster plasmonic resonator for immunological detection of hepatitis B virus. ACS Nano 6(9):7634–7643
Driskell JD et al (2011) One-step assay for detecting influenza virus using dynamic light scattering and gold nanoparticles. Analyst 136(15):3083–3090
Flores CO et al (2011) Statistical structure of host–phage interactions. Proc Natl Acad Sci U S A 108(28):E288–E297
Ganbold E-O et al (2012) Aggregation effects of gold nanoparticles for single-base mismatch detection in influenza A (H1N1) DNA sequences using fluorescence and Raman measurements. Colloids Surf B: Biointerfaces 93:148–153
Gerhardt M et al (2005) In-depth, longitudinal analysis of viral quasispecies from an individual triply infected with late-stage human immunodeficiency virus type 1, using a multiple PCR primer approach. J Virol 79(13):8249–8261
Glynou K et al (2003) Oligonucleotide-functionalized gold nanoparticles as probes in a dry-reagent strip biosensor for DNA analysis by hybridization. Anal Chem 75(16):4155–4160
Griffin J et al (2009) Size-and distance-dependent nanoparticle surface-energy transfer (NSET) method for selective sensing of hepatitis C virus RNA. Chemistry 15(2):342–351
Griffiths C, Drews SJ, Marchant DJ (2017) Respiratory syncytial virus: infection, detection, and new options for prevention and treatment. Clin Microbiol Rev 30(1):277–319
Guo F et al (2010) Valve-based microfluidic device for droplet on-demand operation and static assay. Appl Phys Lett 97(23):233701
Hong SC et al (2015) Continuous aerosol size separator using inertial microfluidics and its application to airborne bacteria and viruses. Lab Chip 15(8):1889–1897
Hong W, Jeong SG, Shim G, Kim DY, Pack SP, Lee CS (2018) Improvement in the reproducibility of a paper-based analytical device (PAD) using stable covalent binding between proteins and cellulose paper. Biotechnol Bioprocess Eng 23(6):686–692
Hu Y, Xu P, Luo J, He H, Du W (2017) Absolute quantification of H5-subtype avian influenza viruses using droplet digital loop-mediated isothermal amplification. Anal Chem 89(1):745–750
Hwang BH, Shin HH, Cha HJ (2017) Optimization of DNA microarray biosensors enables rapid and sensitive detection. Biotechnol Bioprocess Eng 22:469–473
Jeong S-G et al (2015) Toward instrument-free digital measurements: a three-dimensional microfluidic device fabricated in a single sheet of paper by double-sided printing and lamination. Lab Chip 15(4):1188–1194
Laderman EI et al (2008) Rapid, sensitive, and specific lateral-flow immunochromatographic point-of-care device for detection of herpes simplex virus type 2-specific immunoglobulin G antibodies in serum and whole blood. Clin Vaccine Immunol 15(1):159–163
Lecellier CH, Dunoyer P, Arar K, Lehmann-Che J, Eyquem S, Himber C, Saïb A, Voinnet O (2005) A cellular microRNA mediates antiviral defense in human cells. Science 308(5721):557–560
Lee CS, Lee SH, Kim YG, Oh MK, Hwang TS, Rhee YW, Song HM, Kim BY, Kim YK, Kim BG (2006) Fabrication of disposable protein chip for simultaneous sample detection. Biotechnol Bioprocess Eng 11:455
Lei K-M et al (2015) A palm-size μNMR relaxometer using a digital microfluidic device and a semiconductor transceiver for chemical/biological diagnosis. Analyst 140(15):5129–5137
Li H, Rothberg LJ (2004) Colorimetric detection of DNA sequences based on electrostatic interactions with unmodified gold nanoparticles. Proc Natl Acad Sci U S A 101(39):14036–14039
Li H et al (2010) High multiplicity infection by HIV-1 in men who have sex with men. PLoS Pathog 6(5)
Li X et al (2012) A fast and sensitive immunoassay of avian influenza virus based on label-free quantum dot probe and lateral flow test strip. Talanta 100:1–6
Link DR et al (2004) Geometrically mediated breakup of drops in microfluidic devices. Phys Rev Lett 92(5):054503
Liu WW, Goodhouse J, Jeon NL, Enquist LW (2008) A microfluidic chamber for analysis of neuron-to-cell spread and axonal transport of an alpha-herpesvirus. PLoS One 3(6)
Loney C, Mottet-Osman G, Roux L, Bhella D (2009) Paramyxovirus ultrastructure and genome packaging: cryo-electron tomography of sendai virus. J Virol 83(16):8191–8197
Lu X et al (2013) A gold nanorods-based fluorescent biosensor for the detection of hepatitis B virus DNA based on fluorescence resonance energy transfer. Analyst 138(2):642–650
Mancuso M et al (2013) Multiplexed colorimetric detection of Kaposi’s sarcoma associated herpesvirus and Bartonella DNA using gold and silver nanoparticles. Nanoscale 5(4):1678–1686
Martin JG et al (2009) Toward an artificial Golgi: redesigning the biological activities of heparan sulfate on a digital microfluidic chip. J Am Chem Soc 131(31):11041–11048
McCrone JT, Woods RJ, Martin ET, Malosh RE, Monto AS, Lauring AS (2018) Stochastic processes constrain the within and between host evolution of influenza virus. elife 7:e35962
Mitchell P (2002) A perspective on protein microarrays. Nature Biotech 20(3):225–229
Munson-McGee JH et al (2018) A virus or more in (nearly) every cell: ubiquitous networks of virus–host interactions in extreme environments. ISME J 12(7):1706–1714
Neng J et al (2013) Surface-enhanced Raman scattering (SERS) detection of multiple viral antigens using magnetic capture of SERS-active nanoparticles. Biosens Bioelectron 41:316–321
Niu X et al (2007) Real-time detection, control, and sorting of microfluidic droplets. Biomicrofluidics 1(4):44101–44101
Niu X et al (2008) Pillar-induced droplet merging in microfluidic circuits. Lab Chip 8(11):1837–1841
Niu X et al (2009) Droplet-based compartmentalization of chemically separated components in two-dimensional separations. Lab Chip (41):6159–6161
Rosi NL, Mirkin CA (2005) Nanostructures in biodiagnostics. Chem Rev 105(4):1547–1562
Roux S et al (2015) Viral dark matter and virus–host interactions resolved from publicly available microbial genomes. elife 4:e08490
Schmitz CH et al (2009) Dropspots: a picoliter array in a microfluidic device. Lab Chip 9(1):44–49
Sebba D, Lastovich AG, Kuroda M, Fallows E, Johnson J, Ahouidi A, Honko AN, Fu H, Nielson R, Carruthers E, Diédhiou C (2018) A point-of-care diagnostic for differentiating Ebola from endemic febrile diseases. Sci Transl Med 10(471):eaat0944
Shawky SM, Bald D, Azzazy HM (2010) Direct detection of unamplified hepatitis C virus RNA using unmodified gold nanoparticles. Clin Biochem 43(13–14):1163–1168
Shemesh J et al (2010) Advanced microfluidic droplet manipulation based on piezoelectric actuation. Biomed Microdevices 12(5):907–914
Sista RS et al (2011) Digital microfluidic platform for multiplexing enzyme assays: implications for lysosomal storage disease screening in newborns. Clin Chem 57(10):1444–1451
Solvas XC, DeMello A (2011) Droplet microfluidics: recent developments and future applications. Chem Commun 47(7):1936–1942
Spackman E et al (2002) Development of a real-time reverse transcriptase PCR assay for type A influenza virus and the avian H5 and H7 hemagglutinin subtypes. J Clin Microbiol 40(9):3256–3260
Sun Y et al (2019) Correction: a specific sequence in the genome of respiratory syncytial virus regulates the generation of copy-back defective viral genomes. PLoS Pathog 15(10)
Swyer I et al (2016) Interfacing digital microfluidics with high-field nuclear magnetic resonance spectroscopy. Lab Chip 16(22):4424–4435
Talapatra A, Rouse R, Hardiman GJP (2002) Protein microarrays: challenges and promises. Pharmacogenomics 3(4):527–536
Toh SY et al (2015) Aptamers as a replacement for antibodies in enzyme-linked immunosorbent assay. Biosens Bioelectron 64:392–403
Tseng Y-T et al (2016) Integrated microfluidic system for rapid detection of influenza H1N1 virus using a sandwich-based aptamer assay. Biosens Bioelectron 82:105–111
Unger M, et al (2000) Integrated elastomer fluidic Lab-on-a-chip-Surface patterning and DNA diagnostics. 288:113–116
Wang X et al (2009) Engineering nanomaterial surfaces for biomedical applications. Exp Biol Med (Maywood) 234(10):1128–1139
Wang X et al (2010) Gold nanorod-based localized surface plasmon resonance biosensor for sensitive detection of hepatitis B virus in buffer, blood serum and plasma. Biosens Bioelectron 26(2):404–410
Wang X et al (2012) Detection of hepatitis B surface antigen by target-induced aggregation monitored by dynamic light scattering. Anal Biochem 428(2):119–125
Warrick JW, Timm A, Swick A, Yin J (2016) Tools for single-cell kinetic analysis of virus-host interactions. PLoS One 11(1):e0145081
Wu J-C et al (2014) Electrophoresis-enhanced detection of deoxyribonucleic acids on a membrane-based lateral flow strip using avian influenza H5 genetic sequence as the model. Sensors (Basel) 14(3):4399–4415
Xu P, Zheng X, Tao Y, Du W (2016) Cross-interface emulsification for generating size-tunable droplets. Anal Chem 88(6):3171–3177
Xue KS et al (2018) Cooperating H3N2 influenza virus variants are not detectable in primary clinical samples. mSphere 3(1):e00552–e00517
Yeh YT, Gulino K, Zhang Y, Sabestien A, Chou TW, Zhou B, Lin Z, Albert I, Lu H, Swaminathan V, Ghedin E (2020) A rapid and label-free platform for virus capture and identification from clinical samples. Proc Natl Acad Sci 117(2):895–901
Zehbe I et al (1997) Sensitive in situ hybridization with catalyzed reporter deposition, streptavidin-Nanogold, and silver acetate autometallography: detection of single-copy human papillomavirus. Am J Pathol 150(5):1553
Zeng Q et al (2012) Multiple homogeneous immunoassays based on a quantum dots–gold nanorods FRET nanoplatform. Chem Commun 48(12):1781–1783
Author information
Authors and Affiliations
Corresponding author
Editor information
Editors and Affiliations
Rights and permissions
Copyright information
© 2021 Springer Nature Singapore Pte Ltd
About this entry
Cite this entry
Ganguly, R., Lee, CS. (2021). Microfluidic and Nanomaterial Approach for Virology. In: Santra, T.S., Tseng, FG. (eds) Handbook of Single Cell Technologies. Springer, Singapore. https://doi.org/10.1007/978-981-10-4857-9_46-1
Download citation
DOI: https://doi.org/10.1007/978-981-10-4857-9_46-1
Received:
Accepted:
Published:
Publisher Name: Springer, Singapore
Print ISBN: 978-981-10-4857-9
Online ISBN: 978-981-10-4857-9
eBook Packages: Springer Reference EngineeringReference Module Computer Science and Engineering