Summary
RNA silencing, including RNA interference, is a novel method of gene regulation and one of the potent host-defense mechanisms against the viruses. In the course of evolution, the viruses have encoded proteins with the potential to suppress the host RNA silencing mechanism as a counterdefense strategy. The virus-encoded RNA silencing suppressors (RSSs) can serve as important biological tools to dissect the detailed RNA silencing pathways and also to evolve the antiviral strategies. Screening and identification of the RSSs are indeed of utmost significance in the field of plant biotechnology. We describe two Green Fluorescent Protein (GFP) reporter-based plant assay systems that rely on two different principles, namely reversal of silencing and enhancement of rolling circle replication (RCR) of geminiviral replicon. These proof-of-concept examples and assay systems could be used to screen various plant, animal, and insect viral ORFs for identification of the RSS activities.
You have full access to this open access chapter, Download protocol PDF
Similar content being viewed by others
Key Words
- RNA silencing suppressor (RSS)
- GFP
- reversal of silencing
- replication-based spot assay
- rolling circle replication (RCR)
1 Introduction
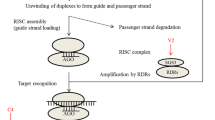
RNA silencing is one of the major adaptive defense responses of the eukaryotic systems (1,2). It is triggered by long dsRNA and targets the homologous ssRNA in a sequence-specific manner. The enzyme Dicer, a prime component of the RNA silencing machinery, degrades the long dsRNA into 20–30-mer short RNAs, known as small interfering RNAs or siRNAs. These siRNAs form a complex with a large repertoire of proteins, known as RISC, which pair with cognate transcripts to slice them up (3–5). In some cases, RNA silencing postviral infection results in a host “recovery” phenotype, in which the newly emerging leaves lack viral symptoms and remain substantially free of virus. Additionally, the RNA silencing processes in response to one mild virus may cross-protect the infected plant against virulent infection by another related virus carrying sequences homologous to the pre-infecting one (6), a phenomenon scripted by the plant virologists long ago in the 1920s. All such phenomena are now generally known as virus-induced gene silencing (VIGS). Moreover, the plant mutants defective in one or several of the RNA silencing pathways are found to show enhanced susceptibility to virus infection (7–10). RNA silencing is also one of the major principles behind “pathogen-derived resistance” (PDR) against viruses. In recent years, the viral ORFs have been used as transgenes of inverted repeats with intervening introns to ensure production of viral siRNAs within the transgenic plants. Such plants mount robust resistance against the challenge viruses harboring the genomic sequences similar to those of the transgenes. These facts essentially advocate the antiviral characteristics of RNA silencing.
However, the viruses have also evolved to overcome this strong host-defense response of RNA silencing and cause severe pathogenic symptoms and diseases. Moreover, there is evidence of synergism during co-infection, where the weak viral symptoms are magnificently aggravated following synergism (11). The virus-resistant transgenic plants based on the RNA silencing principle could also succumb to viral infection as a result of synergistic co-infection. There are certain viral proteins that neither directly support the replication of the viral genome nor fall under classical pathogenicity factors, but are found to be important for the establishment of disease (12). The transgenic plants expressing such viral proteins are more susceptible to viral infection than the wild-type plants; and the viruses defective for such proteins are either less virulent than the wild-type counterparts or are completely disabled in growth by host RNA silencing processes (13–15). These facts cumulatively underscore the point that viruses encode a few proteins to counteract the host RNA silencing and these proteins confer pathogenicity to the viruses. These proteins are known as RNA silencing suppressors (RSSs). The potyvirus HC-Pro protein was the first RSS to be identified (16) and then five more RSSs were reported in 1999 (17). Since then a large number of suppressors (about 45) have been identified from plant, animal, as well as insect viruses (18).
The RSSs have the potential to act across the kingdom of life, i.e., the animal virus-encoded RSS can suppress the plant RNA silencing pathways, and vice versa. However, besides the commonality that they can suppress RNA silencing, they differ in evolution, origin, sequence, protein 3D-structure, and modes of action. Thus, identification and characterization of suppressors from individual virus groups are needed. The RSSs can also be used to decipher the detailed pathway of RNA silencing as different suppressors act at different biochemical steps of RNA silencing. The screening of virus ORFs for RSS activity is also essential to design virus resistance strategy, as the RSS encoding virus would not easily succumb to RNA silencing strategy. The identified RSS can also be used to enhance transgenic protein production, as posttranscriptional gene silencing (PTGS) has been found to be one of the limiting factors for transgene expression (19).
In principle, most, if not all, of the viruses might encode RSSs, but only a few have been identified. The availability of the reliable screening system or assay protocol is one of the limitations for the identification of RSSs. Here we primarily discuss two assay systems that we have developed in our laboratory, the applications of which have allowed us to successfully screen many viral RSSs (Table 1).
1.1 Reversal of Reporter Transgene Silencing Assay
The reversal of silencing assay is a widely used reporter-based assay system. It is based upon the principle that suppression of RNA silencing would lead to the expression of silenced reporter gene, such as the GFP transgene in this particular case. Here we use the GFP-silenced transgenic tobacco plant, cv. xanthi, where the GFP gene is chromosomally integrated but silent in expression due to posttranscriptional gene silencing. Various ORFs of the viral origin are allowed to ectopically express in the leaves of the silent plant following agro-inoculation. In some cases the ectopic expression will lead to reversion of silenced phenotype, i.e., GFP expression. The reversion of expression can also be monitored by Northern analyses. The candidate viral ORFs that lead to the eventual reversion of GFP expression could be adjudicated as the suppressors of RNA silencing.
The RSSs identified by this assay primarily have the potential to revert silencing that is pre-established. In order to identify the suppressor that interferes at the initiation or establishment of RNA silencing, we developed another assay system, namely replication-based spot assay.
1.2 Replication-Based Spot Assay
The method is primarily based upon the principle that RNA silencing restricts the viral genome replication by targeting the viral RNA transcript and/or RNA genome and the RSSs, which suppress RNA silencing, should enhance replication when provided in adequate amounts in trans.
We have made use of the characteristics of rolling circle replication (RCR) of a geminivirus-based amplicon (VA) that replicates like an independent episome in plants (20,21). The left and right borders of a standard plant transformation vector (e.g., pCAMBIA or pBI121) are joined by a DNA segment consisting essentially of the following three components. These include the replicon part of the DNA-A component of Mung bean yellow mosaic India virus (MYMIV) containing the cis elements of replication origin (CR) along with three ORFs, namely Rep, AC2, and AC3; the transcription unit of GFP; and the replication origin alone (CR) (Figs. 1a and 1b). When the resultant plasmid (say, pCAMBIA-VA/GFP) is introduced in the leaves of N. xanthi by agro-inoculation, the DNA spaced between the two CRs excise out to form a replicating episomal circle. The Rep protein of MYMIV essentially mediates this excision and subsequent initiation of replication (RCR). The amount of DNA replication can be measured by three independent means, as mentioned later. The episomal replication and consequent GFP fluorescence are detected within a week of agro-inoculation at the inoculated spot, but the fluorescence disappears at two weeks’ postinoculation due to silencing of the GFP transcripts and short life-span of the GFP protein. The silencing effect is even more potent when the AC2 of VA is mutated, confirming the nature of AC2 as an RNA silencing suppressor. This disappearance, however, can be restored if the RNAi-suppressor-expressing construct could be co-introduced along with the pCAMBIA-VA/GFP construct using the Agrobacterial route. In the presence of the suppressor, more episomal replication along with the stable GFP fluorescence is observed even at four weeks’ postinoculation.
MYMIV-based viral amplicon with reporter gene GFP (VA/GFP). (a) The map of MYMIV-DNA “A” genome. The map shown on the left side of the panel depicts the positions of different ORFs and the location of CR-AC3 (functional replicon) used for the study. (b) Map showing the VA vector and its cloning sites. The engineered viral insert containing GFP as a marker was cloned in the pCAMBIA1391Z background. The circular episome that formed in the plant leaves due to replication-release following agro-inoculation is shown schematically using a curved arrow.
We screened innumerable ORFs from various viruses, namely Mung bean yellow mosaic India virus (MYMIV), a huge number of other geminiviruses, Flock house virus (FHV), Severe acute respiratory syndrome (SARS) virus, etc., using the above-mentioned assays. The ORFs that were adjudicated as RSSs are shown in Table 1.
2 Materials
2.1 Plasmids, Culture, Medium, and Buffers
-
Plasmids: pCAMBIA1391Z (CAMBIA, Australia), pBI121 (Clontech)
-
Culture: Agrobacterium tumefaciens (LBA4404)
-
Plant: Nicotiana tabaccum var. xanthi
-
Genes: Mungbean yellow mosaic India virus ORF AC2 and other AC2 homologues from geminiviruses
-
Flock house virus ORF-B2
-
Severe acute respiratory syndrome virus ORF 3a, 7a, and N
-
Medium: Yeast extract mannitol broth/agar
-
Yeast extract: 0.4 g/L
-
Mannitol: 10.0 g/L
-
NaCl: 0.1 g/L
-
MgSO4.7H2O: 0.2 g/L
-
K2HPO4: 0.5 g/L
-
pH: 7.0
-
Agar: 15 g/L
-
CTAB (Hexadecyltrimethylammonium bromide) buffer: 55 mM of CTAB; 1.4 M of NaCl; 20 mM of EDTA; 100 mM of Tris, pH 8.0; 128 mM of β-mercaptoethanol.
2.2 RNA Gel
2.2.1 For mRNA:
-
1.
Dissolve 1 g of agarose in 84 mL of DEPC-treated autoclaved water. Cool to 65 °C and add 10 mL of 10X MOPS, 4.5 mL of 37% w/v formaldehyde, and 7 μL of 10 mg/mL EtBr.
-
2.
10X MOPS: 0.4 M MOPS, 0.1 M Sodium acetate, 10 mM Na EDTA, pH: 7.2
2.2.2 For small RNA:
-
1.
Gel composition: For 7 M of urea-15% polyacrylamide gel (20 mL): 40% Acrylamide: bis-acrylamide (19:1): (7.5 mL) Urea: (4.2 g) 10% APS: (200 μL) 10XTBE: (1000 μL) TEMED: (10 μL) DEPC-treated water: to make up volume to 20 mL.
-
2.
Running buffer: 0.5X TBE.
-
3.
RNA loading buffer: (2X): Formamide: 720 μL, 10X MOPS: 160 μL, 37% formaldehyde: 260 μL, DEPC-treated water: 180 μL, 80% glycerol: 100 μL, bromophenol blue: 80 μL (saturated solution).
2.3 Northern Analysis
2.3.1 Prehybridization buffer (1,000 mL):
-
1.
250 mL 20X SSC, 100 mL 50% dextran sulphate, 50 mL 1.0 M Sodium phosphate pH 7.2, 100 mL 50X Denhardt’s Solution. 0.5 M EDTA: 5 mL 20% SDS: 20 mL Distilled water: 475 mL (Divided into 50-mL aliquots and stored at mbox–20 °C). In case of siRNA hybridization, also add 30% v/v formamide in above prehybridization buffer.
-
2.
Hybridization buffer: prehybridization buffer + radiolabeled probe.
2.4 PCR Primers to Quantitate Episomal Replication
-
AC1 R: AGA AGC TTC TAT GCG TCG TTG GCA GAT TG
-
GFP F: GCT CTA GAC CAT GGC AAG TAA AGG AGA AGA ACT T
3 Methods
The viral ORFs to be screened for RSS activity should be cloned in binary vectors like pBI121, under a strong promoter, such as the 35S promoter, so that strong transient expression of the gene can be obtained in the subsequent assays. The constructs are transformed into laboratory strains of agrobacteria, such as Agrobacterium tumefaciens LBA4404, as the assays are in planta-based.
3.1 Preparation of Agrobacterium Culture for Infiltration
Agrobacterium LBA4404 strain harboring empty vector (pBI121) with or without the viral ORFs is streaked onto LB agar plate with selection antibiotic, kanamycin (50 mg/mL) for the plasmid and streptomycin (25 mg/mL) for the agro-bacterial strain, and incubated at 30 °C. The single colony thus obtained is inoculated into 5 mL of YEM medium as primary culture and grown at 30 °C, 200 rpm, overnight. About 2% of the primary inoculum is then used to inoculate 50 mL of the YEM broth in a 500-mL Erlenmeyer flask and grown at 30 °C, 200 rpm, until the O.D.600nm reaches 0.8–1.0 (see Note 1). All the inoculations are carried out under aseptic conditions.
3.2 Screening of Suppressor Activity by Reversal of Silencing Assay
The outline of the experiment is sketched in Fig. 2. The method is based on the reversion of reporter gene GFP expression from the pre-established silenced state in the presence of viral ORFs. The GFP-silenced and wild-type leaves, under U.V. (~311 nm) exposure, fluoresce red light (~680 nm) due to the presence of chlorophyll, while the GFP expression leads to green fluorescence at ~510 nm. The molecular analyses involved probing the mRNA and siRNA levels of GFP in the presence and absence of the viral ORFs.
Diagrammatic representation of reversal of silencing assay. (a) GFP-silenced transgenic plant showed red fluorescence under U.V. due to chlorophyll. (b) The transgenic plant was agro-infiltrated with test ORFs. (c) GFP fluorescence at the zone of infiltration observed after 8–9 days’ postinfiltration. (d) No reversion of silencing if infiltration was carried out with nonsuppressor construct.
-
1.
The GFP-silenced Nicotiana tabacum cv. xanthi can be made by Agrobacterium-mediated transformation of the wild-type Nicotiana plant using the pBI121-GFP constructs. A transgenic event is selected in which the chromosomally integrated GFP gene is silent in expression in Nicotiana xanthi. The transgenics are screened for loss of fluorescence following UV irradiation on leaves, while the presence of the GFP gene is confirmed by both PCR and Southern analyses. We also confirm the silencing by the very low level of GFP-mRNA and the high level of GFP-siRNA in the Northern blots of the RNAs isolated from the silent plants. The young leaves of the transgenic plant are used for the screening assay.
-
2.
Take up the homogeneous culture suspension of Agrobacterium constructs (obtained in Section 3.1) in a 5-mL needleless syringe. Infiltrate the selected young leaf using the culture suspension and by generating a vacuum with the help of a finger on the dorsal side of the leaf and mouth of the syringe on the ventral side. As the vacuum is created, push the plunger of the syringe to transfer the culture into the leaf through the stomata pores without detachment of the leaf from the plant (see Notes 2 and 3). Label each agro-infiltrated leaf for the construct used for infiltration. A plant should not be infiltrated for more than two ORFs for best results (see Note 4).
-
3.
Pluck the inoculated leaves from the plant starting from 4 days’ postinfiltration (d.p.i.) for fluorescence and biochemical analyses. The incubation period is required for the optimum expression and suppression activity of the viral ORF (see Note 5).
-
4.
Visualize the leaves under UV for fluorescence. In case of an empty vector (pBI121), SARS 3a and N, the infiltrated zone will appear red at any time postinfiltration, while the MYMIV-AC2, FHV-B2, and SARS-7a infiltrated zones will fluoresce green at about 8 d.p.i. (Fig. 2). Use the leaves for total RNA and small RNA isolation. There are various common laboratory methods for this, and an example is given below.
-
5.
Isolate total RNA from plant tissue by the TRIzol reagent method (Gibco BRL) (see Note 6).
-
a.
Homogenize plant tissue (see Note 7) in liquid nitrogen and add TRIzol reagent to the homogenate (1 mL per 100 mg of tissue). Incubate at room temperature for 5 min.
-
b.
Add 200 μL of chloroform per 1 mL of TRIzol reagent to the mixture, mix by vigorously shaking for 15 sec, and incubate again at room temperature for 2–3 min. Centrifuge the homogenate at 12,000 x g for 15 min at 4 °C.
-
c.
Pipette out the supernatant into a fresh RNase-free 1.5-mL Eppendorf tube while the pellet containing cell debris is discarded. Add about 0.7 volume of isopropanol to the supernatant, centrifuge to precipitate the RNA, and pellet down at 10,000 x g for 10 min.
-
d.
Wash the pellet with 75% ethanol made in DEPC-treated water, air-dry the RNA, and dissolve in DEPC-treated water (see Notes 8 and 9).
-
a.
-
6.
The total RNA is further processed for small RNA isolation as follows.
-
a.
Heat the RNA samples at 65 °C for 10 min to disrupt the association between larger- and smaller-size RNA molecules and also to facilitate dissolving of the pellet.
-
b.
Place the dissolved RNA immediately on ice and add PEG (MW 8,000) to a final concentration of 5% and NaCl to a final concentration of 0.5 M. Mix and keep in ice for 30 min (see Note 10).
-
c.
Centrifuge at 10,000 x g for 10 min to pellet down the high molecular weight nucleic acids.
-
d.
Collect the supernatant (which contains mainly the small RNAs, namely siRNA, miRNA, degraded RNA, tRNA, and rRNA, etc.), add three volumes of absolute ethanol to it, mix well, and keep at –20 °C for 2 h.
-
e.
Centrifuge at 10,000 x g for 10 min. The pellet thus obtained contains all forms of small RNAs.
-
f.
Wash the pellet with DEPC-treated 75% ethanol, air-dry, and dissolve in DEPC-treated water before using.
-
a.
-
7.
Northern analysis to detect GFP mRNA and siRNA level in the infiltrated leaves is carried out as follows.
-
a.
mRNA level of GFP:
-
i.
Size-fractionate total RNA (as obtained in step 5d) on 1.0% formaldehyde denaturing agarose gel in 1X MOPS running buffer.
-
ii.
After completion of the run, rinse the gel in DEPC-treated water to remove the formaldehyde from the gel and saturate with 10X SSC for 45 min.
-
iii.
Transfer RNA to nylon membrane (Hybond-N, Amersham) by capillary blotting in 10X SSC buffer.
-
iv.
Cross-link the RNA to membrane using a UV cross-linker (Stratagene).
-
v.
Carry out prehybridization and hybridization for the RNA blot following standard protocols (see Notes 12 and 13).
-
vi.
Following hybridization, wash the membrane in 2X SSC, 2% SDS, twice (15 min each time) at room temperature and then twice with 1X SSC, 0.2% SDS, at 65 °C for 25 min each time.
-
vii.
Check the membrane regularly for counts using a Gieger–Müller counter, and make the final washes in 0.1X SSC for 15 min at 60 °C.
-
viii.
Gently wipe the membrane to remove extra buffer and place on an equal-sized Whatman filter sheet (see Note 14).
-
ix.
Wrap the membrane in Saran Wrap and expose to the storage phosphor screen for a desired period of time. The presence or absence of the band in the membrane is visualized by scanning with a phosphorimager scanner. We use Typhoon 9213 and then analyze the intensity of the autoradiographic band using the Image quanta TL program.
-
i.
-
b.
siRNA level of GFP:
-
i.
Resolve the low molecular weight RNA containing siRNA (as obtained in step 6f) in 15% acrylamide-7 M urea gel in 0.5X TBE by overnight electrophoresis at 2.5 V/cm (see Note 11).
-
ii.
Transfer the nucleic acids to a nylon membrane by electro-blotting in 0.5X TBE transfer buffer at 20 V for 40 min.
-
iii.
Cross-link in a UV cross-linker as before.
-
iv.
Perform prehybridization in 30% formaldehyde hybridization buffer for 2 h with mild agitation at 42 °C followed by hybridization with the GFP probe for 14–16 h (see Note 12).
-
v.
Following hybridization, wash the blot at 40 °C with 0.5X SSC, 0.1% SDS for 30 min, and analyze in a phosphorimager (see Notes 13 and 14).
-
i.
The GFP-mRNA levels are generally higher in case of RNA isolated from leaves infiltrated with FHV-B2, MYMIV-AC2, and SARS-7a ORFs but are below a detectable level for the empty vector, SARS-3a, and SARS-N ORFs. However, the GFP-siRNA level is higher for the empty vector, SARS-3a, and SARS-N and lower with FHV-B2, MYMIV-AC2, and SARS-7a. These findings corroborate well with the fluorescence data, thereby proving the RSS activity of FHV-B2, MYMIV-AC2, and SARS-7a. The reversal of the silencing assay is indeed a very easy and reliable method to screen and identify various viral ORFs with RSS activity.
-
a.
3.3 Replication-Based Spot Assay
The DNA-A component of MYMIV replicates in the RCR mode from its replication origin CR (0.2 kb), and its essential replicon is a portion of DNA-A that spans the CR-AC3 region (1.6 kb) as shown in Fig. 1a (20,22). A GFP transcription unit of 1.4 kb consisting of 35S promoter, GFP ORF, and the transcription terminator was prepared keeping the MYMIV-based replicon and the CR at the flanking ends of the transcription unit as shown in Fig. 1b. The resulting construct, when introduced in plants, generated an independently replicating episomal circle of 3.2 kb (Fig. 3a) (20,21). The circle could be detected as a 3.2-kb Southern band and could be distinguished from the unreplicated DNA that would reveal as a ~4 kb band (Fig. 3c). The presence of the circle could also be confirmed from a PCR band of 1.6 kb using a set of divergent primers as mentioned later. There won’t be any PCR amplification from the unreplicated DNA using the same set of primers (Figs. 3a, 3d). At the initial phase of episomal replication, the GFP accumulates showing the green fluorescence in the leaves harboring the replicated DNA. However, around 12 d.p.i., the GFP fluorescence starts fading, showing the signs of GFP silencing (Fig. 3b). At three weeks’ postreplication, the GFP fluorescence is almost lost. Thus, the GFP serves as a marker of the initial phase of replication, as its expression would be directly proportional to the replication of the episome by RCR. The primary plasmid construct, which is introduced in the plant using agro-inoculation, is named VA/GFP and its backbone is pCAMBIA 1391Z (Fig. 3a).
The Southern and PCR-based analysis to distinguish between the replicated and unreplicated forms of pCAMBIA/VA/GFP DNA. (a) The schematics of the experiment. Arrows represent the primers designed for the PCR and the indicated ScaI restriction sites are used for Southern analysis. (b) Diagrammatic representation of agro-infiltration of VA/GFP (upper panel) and time kinetics of GFP fluorescence in the infiltrated zone in tobacco leaf at 0, 3, 10, 15, and 20 d.p.i. (lower panel). Fluorescence was observed by using a low-magnification objective LWD 20C0.40 ph1 of an inverted fluorescent microscope (Nikon, Eclipse TE2000U). (c) Southern analysis of ScaI digested genomic DNA isolated from agro-inoculated tobacco. (d) Detection of circular amplicon by PCR. The plasmid DNA of the amplicon (Cam-VA/GFP), isolated from E. coli, was taken as a negative control for PCR using the primers mentioned in Section 2.4 (lane 1). The genomic DNA isolated from Cam/VA infiltrated leaf after 7 d.p.i., without (lane 2) and with DpnI digestion (lane 3), was taken as a template of the PCR for 21 cycles. The lower panel shows the respective amplification of actin as control.
On the other hand, if the VA/GFP construct is co-inoculated with one expressing the suppressor of RNA silencing, the GFP fluorescence remains stable for more than 30 d.p.i. The suppressors not only stabilize the GFP fluorescence but also enhance the amount of episomal replication (Figs. 4a, 4b).
Enhanced replication of MYMIV-based viral amplicon (VA) in the presence of RNA silencing suppressors. (a) GFP expression in the leaves of Nicotiana infiltrated with AC2 mutant amplicon plus empty pBI121 vector, the amplicon plus empty pBI121 vector, AC2 mutant amplicon plus pBI121-RNA silencing suppressors (AC2, FHV-B2, SARS-7a) at 18 days’ postinfiltration. (b) The PCR-based analysis to determine the enhancement of the replication in presence of AC2, FHV-B2, and SARS-7a. The right panel shows the respective amplification of actin as a loading control.
The episomal elements of the VA/GFP construct can be cloned in a standard plant transformation vector. We cloned the amplicon element at the HindIII-EcoR1 restriction sites of the pCAMBIA vector (Fig. 1b or 3a) and named it viral amplicon (VA) because of its competence to release the episomally replicating circle. We introduced the pCAMBIA-VA/GFP construct into Agrobacterium strain LBA4404 and grew it in a similar manner as described in Section 3.1 (see Note 1). The screening for RSS activity can then be carried out using the following procedures:
-
1.
Mix VA/GFP and either the test viral ORF/empty vector (we used the same ORF that was tested with reversal of silencing assay) or the empty vector carrying culture suspensions of Agrobacteria at 1:1 ratio and agro-infiltrate onto wild-type Nicotiana tabacum cv. xanthi leaves following the procedures described in Section 3.2 (see Notes 2, 3, and 4).
-
2.
Pluck the leaves after 18 d.p.i. and observe under UV for the level of GFP expression (Fig. 4) (see Note 5).
-
3.
Extract the genomic DNA from the infiltrated zones using the following steps:
-
a.
Grind the plant material in liquid nitrogen (2 g of fresh tissue or 0.5 g of dried tissue) to fine powder and add 5 mL of CTAB buffer to make a slurry (see Note 7).
-
b.
Transfer the slurry to an autoclaved propylene tube and incubate at 60 °C in a shaking water bath (100 rpm) for 35 min.
-
c.
Add an equal volume of chloroform–isoamyl alcohol (24:1) and mix gently.
-
d.
Centrifuge at 10,000 x g for 5 min at room temperature.
-
e.
Carefully remove the upper aqueous layer with a wide-bore pipette. Add 0.7 vol. of isopropanol and mix gently by inversion.
-
f.
Centrifuge the sample at 10,000 x g for 10 min at room temperature to precipitate the nucleic acids.
-
g.
Discard the supernatant and wash the pellet with 80% ethanol two to three times.
-
h.
Air-dry the pellet until the ethanol disappears (about 30 min). Dissolve in 0.5–1 mL of TE buffer and collect in an autoclaved 1.5-mL Eppendorf tube.
-
i.
After dissolving the nucleic acids, add 5 μL of RNase A (10 mg/mL) and incubate at 37 °C for 30 min.
-
j.
Add 1/2 volof Tris-saturated phenol and 1/2 volof chloroform–isoamyl alcohol (24:1), mix, and centrifuge at 10,000 x g for 10 min at room temperature.
-
k.
Transfer the upper aqueous layer to a fresh 1.5-mL Eppendorf tube, mix an equal volume of chloroform–isoamyl alcohol (24:1), and centrifuge at 10,000 x g for 10 min at room temperature. Repeat this step twice.
-
l.
Finally, take out the upper aqueous phase and precipitate the DNA with 1/10 vol of 3 M sodium acetate (pH 5.2) and 2 vol of ethanol. Centrifuge at 10,000 x g for 10 min at 4 °C.
-
m.
Wash the pellet with 70% ethanol, air-dry, and dissolve in 0.3 mL of sterile distilled water (see Notes 8 and 9).
-
a.
-
4.
Use the isolated DNA for the PCR-based replication analysis. Design two primers, GFP F and AL1 R, such that a PCR amplification product of 1.6 kb will result if the circular episome template is present. The parental DNA template of around 11 kb would not produce any PCR product because of the nonprocessive characteristics of the thermophilic DNA polymerase (lane 1, Fig. 3d). Perform 50-μL PCR for 21 cycles using 0.5 mM dNTPs at 54 °C annealing temperature and 72 °C extension for 2 min (see Note 14). Test that the DpnI digestion of the DNA template prior to PCR does not make any difference, indicating the replicative nature of the episomal DNA template.
-
5.
Analyze the PCR product along with a 1-kb marker ladder in a 1% agarose gel with 1X TBE running buffer and observe in a UV transilluminator for EtBr-stained bands.
In all the samples, the 1.6-kb bands are observed. However, the band intensity will vary between the samples. The minimum intensity of band resulted with VA+empty vector, and a higher intensity was observed with VA+RSS. For internal control, amplify actin using specific primers in each PCR (Fig. 4b). In conclusion, these assays should serve as simple and rapid strategies for the screening and identification of the viral ORFs with the RSS activity.
4 Notes
-
1.
To screen RNA silencing suppressor activity, the Agrobacterium harboring viral ORF should be grown to an optical density of 0.8–1.0 at 600 nm.
-
2.
Agro-infiltration should be carried out gently in order to avoid any mechanical injury to the leaf.
-
3.
The plant to be agro-infiltrated should be young, about 6–8 weeks old.
-
4.
Do not infiltrate a plant for more than two ORFs, as it would otherwise lead to false-positive results.
-
5.
Optimize the incubation period between infiltration and plucking of leaf for the test ORFs in order to obtain the best results.
-
6.
Ensure that the equipments and reagents to be used for RNA isolation are properly autoclaved and RNase-free.
-
7.
Isolate DNA or RNA from the infiltrated zone and not the entire leaf.
-
8.
Store DNA and RNA samples properly at –20 °C and –80 °C, respectively.
-
9.
Avoid repeated freezing and thawing of the samples by storing them in small aliquots.
-
10.
In PEG-NaCl precipitation for small and large RNA fractionation, do not incubate the samples for more than 30 min.
-
11.
The urea-PAGE should be carried out at room temperature (and not in the cold), as it otherwise leads to crystallization of urea.
-
12.
To ensure efficient probe preparation, purify the probe and check by scintillation counting before addition.
-
13.
Perform prehybridization, hybridization, and washing for the prescribed time in order to obtain clean results.
-
14.
PCR should be carried out for a restricted number of cycles such that the product does not reach saturation level and differences in the PCR product band intensity can be analyzed.
References
1. Hamilton, W. D. O., and Baulcombe, D. C. (1989). Infectious RNA produced by in vitro transcription of a full-length tobacco rattle virus RNA-1 cDNA J. Gen. Virol. 70, 963–968.
2. Ratcliff, F., Harrison, B. D., and Baulcombe, D. C. (1997). A similarity between viral defense and gene silencing in plants Science 276, 1558–1560.
3. Agrawal, N., Dasaradhi, P. V. N., Mohmmed, A., Malhotra, P., Bhatnagar, R. K., and Mukherjee, S. K. (2003). RNA interference: Biology, mechanism, and applications Microbiol. Mol. Biol. Rev. 67, 657–685.
4. Cerutti, H. (2003). RNA interference: Traveling in the cell and gaining functions? Trends Genet. 19, 39–46.
5. Hannon, G. J. (2002). RNA interference Nature 418, 244–251.
6. Ratcliff, F. G., MacFarlane, S. A., and Baulcombe, D. C. (1999). Gene silencing without DNA: RNA-mediated cross-protection between viruses Plant Cell 11, 1207–1216.
7. Dalmay, T., Hamilton, A. J., Rudd, S., Angell, S., and Baulcombe, D. C. (2000). An RNA-dependent RNA polymerase gene in Arabidopsis is required for posttranscriptional gene silencing mediated by a transgene but not by a virus Cell 101, 543–553.
8. Mourrain, P., Beclin, C., Elmayan, T., et al. (2000). Arabidopsis SGS2 and SGS3 genes are required for posttranscriptional gene silencing and natural virus resistance Cell 101, 533–542.
9. Boutet, S., Vazquez, F., Liu, J., et al. (2003). Arabidopsis HEN1: A genetic link between endogenous miRNA controlling development and siRNA controlling transgene silencing and virus resistance Curr. Biol. 13, 843–848.
10. Wilkins, C., Dishongh, R., Moore, S. C., Whitt, M. A., Chow, M., and Machaca, K. (2005). RNA interference is an antiviral defence mechanism in Caenorhabditis elegans Nature 436, 1044–1047.
11. Vance, V. B. (1991). Replication of potato virus X RNA is altered in coinfections with potato virus Y Virology 182, 486–494.
12. Pruss, G., Ge, X., Shi, X. M., Carrington, J. C., and Vance, V. B. (1997). Plant viral synergism: The potyviral genome encodes a broad-range pathogenicity enhancer that transactivates replication of heterologous viruses Plant Cell 9, 859–868.
13. Chellappan, P., Vanitharani, R., and Fauquet, C. M. (2005). MicroRNA-binding viral protein interferes with Arabidopsis development Proc. Natl. Acad. Sci. USA 102, 10381–10386.
14. Brigneti, G., Voinnet, O., Li, W. X., Ji, L. H., Ding, S. H., and Baulcombe, D. C. (1998). Viral pathogenicity determinants are suppressors of transgene silencing in Nicotiana benthamiana EMBO J. 17, 6739–6746.
15. Li, H., Li, W. X., and Ding, S. W. (2004). Induction and suppression of RNA silencing by an animal virus Science 296, 1319–1321.
16. Anandalakshmi, R., Pruss, G. J., Ge, X., Marathe, R., Mallory, A. C., Smith, T. H., and Vance, V. B. (1998). A viral suppressor of gene silencing in plants Proc. Natl. Acad. Sci. USA 95, 13079–13084.
17. Voinnet, O., Pinto, Y. M., and Baulcombe, D. C. (1999). Suppression of gene silencing: A general strategy used by diverse DNA and RNA viruses of plants Proc. Natl. Acad. Sci. USA 96, 14147–14152.
18. Li, H., and Ding, S.W. (2006). Virus counterdefense: Diverse strategies for evading the RNA-silencing immunity Ann. Rev. Microbiol. 60, 503–531.
19. Johansen, L. K., and Carrington, J. C. (2001). Silencing on the spot. Induction and suppression of RNA silencing in the Agrobacterium-mediated transient expression system Plant Physiol. 126, 930–938.
20. Roy Choudhury, N., Malik, P. S., Singh, D. K., Islam, M. N., Kaliappan, K., and Mukherjee, S. K. (2006). The oligomeric Rep protein of Mung bean yellow mosaic India virus (MYMIV) is a likely replicative helicase Nucleic Acids Res. 34, 6362–6377.
21. Singh, D. K., Islam, M. N., Roy Choudhury, N., Karjee, S., and Mukherjee, S. K. (2007). The 32 kDa subunit of replication protein A (RPA) participates in the DNA replication of Mung bean yellow mosaic India virus (MYMIV) by interacting with the viral Rep protein Nucleic Acids Res. 35, 755–770.
22. Pant, V., Gupta, D., Choudhury, N. R., Malathi, V. G., Varma, A., and Mukherjee, S. K. (2001). Molecular characterization of the Rep protein of the blackgram isolate of Indian mung bean yellow mosaic virus J. Gen. Virol. 82, 2559–2567.
Acknowledgments
The authors would like to thank Dr. Nirupam Roy Choudhury, Dr. Dharmendra K. Singh, and Mrs. Subhra (Datta) Mukhopadhyay of ICGEB, New Delhi for their helpful input, and Dr. Ranjit DasGupta of the University of Wisconsin, Madison, for providing the FHV-RNA1 genome. The SARS-3a, 7a, and N genes were kind gifts from the laboratory of Dr. Sunil K. Lal of ICGEB, New Delhi. The financial assistance of CSIR (India) to S. K. is duly acknowledged.
Author information
Authors and Affiliations
Editor information
Editors and Affiliations
Rights and permissions
Copyright information
© 2008 Humana Press, a part of Springer Science+Business Media, LLC
About this protocol
Cite this protocol
Karjee, S., Islam, M.N., Mukherjee, S.K. (2008). Screening and Identification of Virus-Encoded RNA Silencing Suppressors. In: Barik, S. (eds) RNAi. Methods in Molecular Biology™, vol 442. Humana Press. https://doi.org/10.1007/978-1-59745-191-8_14
Download citation
DOI: https://doi.org/10.1007/978-1-59745-191-8_14
Publisher Name: Humana Press
Print ISBN: 978-1-58829-874-4
Online ISBN: 978-1-59745-191-8
eBook Packages: Springer Protocols