Abstract
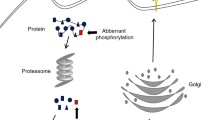
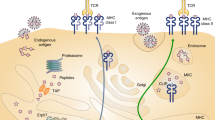
Phosphorylation is one of the most important forms of posttranslational modification. Dysregulation of phosphorylation is implicated in tumorigenesis, with cancerous signaling pathways activated by kinases. For immunotherapy with neoantigen-based peptides, phosphopeptides derived from aberrantly phosphorylated proteins presented by major histocompatibility complex (MHC) are promising candidates due to their specificity to elicit cytotoxic T-cell responses. Unlike other MHC peptides, phosphorylated MHC peptides cannot be predicted from DNA sequences, and their identification relies on the direct detection of phosphopeptides using mass spectrometry (MS). For MS detection, it is extremely important to enrich phosphorylated peptides from the complex repertoire of MHC peptides. Herein, we describe the combined use of immobilized metal affinity chromatography and titanium dioxide nanoparticles for phosphopeptides enrichment from immunopeptidome.
Similar content being viewed by others
Key words
1 Introduction
Protein phosphorylation by kinases and dephosphorylation by phosphatases are key components of protein signaling in cellular networks [1]. It was estimated that phosphorylation affects around 30% of the proteome and regulates many cellular processes [2]. Dysregulation of protein phosphorylation has been found to be a unique feature of cancer [3], with different phosphorylation pathways observed to be activated in tumors. Quantitative analysis of altered phosphorylation will provide unique insights into cancer, the identification of cancer-specific biomarkers and discovery of new targets of cancer therapy [4,5,6]. With the latest advances in mass spectrometry (MS) techniques, especially enhanced scanning speed and detection sensitivity, phosphoproteomics has revealed tumor-associated signatures from various samples, such as cancer cell lines [7], cancer stem cells [8], patient-derived xenograft [9], and tumor tissues [10]. Phosphorylation is more effective in triggering an immune response from cytotoxic T cells than unmodified peptides [6, 11]. Identification of phosphorylated peptides bound to the major histocompatibility complex (MHC) is important in the development of cancer immunotherapy with neoantigens. Phosphorylated MHC peptides have been detected directly from LC-MS/MS analysis of whole immunopeptidome isolated from cells or tissues with immunoprecipitation [12, 13]. However, the low abundance of phosphorylated MHC peptides in comparison to their non-modified counterparts makes their detection and identification challenging. Thus, enrichment of phosphopeptides prior to LC-MS/MS analysis is crucial to increase the specificity and accuracy in identification of phosphorylated MHC peptides.
There are several published protocols for phosphopeptide enrichment that are compatible with downstream MS analyses. The majority of them are based on immobilized metal affinity chromatography (IMAC) or metal oxide affinity chromatography (MOAC) [14]. Iron chelated to nitrilotriacetic was the first IMAC exploited for phosphopeptide enrichment. This was superseded by metal oxide particles and Ti4+ IMAC [15,16,17]. TiO2 and Ti4+ IMAC enrichment methods have a higher affinity for phosphopeptides than that of Fe3+-based IMAC. Unfortunately, the Ti4+ IMAC used in their report is not available to the public. Most large-scale studies still use TiO2 for the enrichment due to its high reproducibility. Recently, improvement in Fe3+ IMAC suggested that nearly 100% specificity for phosphopeptides could be achieved [18]. The literature suggests that the coverage of the phosphoproteome was primarily determined by the metal ion used for enrichment. Some reports show that different methods provided varied immunopeptidome coverage [19].
In this protocol, we present a complementary enrichment of phosphorylated MHC peptides with Fe3+ IMAC and TiO2 nanoparticles. This strategy provides a mechanism to enhance phosphopeptide coverage without sacrificing sensitivity and specificity. The proposed method was evaluated by analyzing immunopeptidome from A549 cell lines, and complementary identification of phosphorylated MHC peptides was achieved (see Table 1).
2 Materials
2.1 Cell Culture
-
1.
Cells: Human lung cancer cell line A549 obtained from the American Type Culture Collection (ATCC).
-
2.
Culture media: Dulbecco’s Modified Eagle’s medium (DMEM) with 10% fetal bovine serum (FBS) and penicillin-streptomycin solution (final concentration of 1000 U/mL and 1 mg/mL, respectively).
2.2 Immuno-precipitation
-
1.
Cell lysis buffer: 200 mM sodium chloride, 20 mM Tris–HCl (pH 7.4), 0.5% CHAPS, 5 mM sodium vanadate, and 5 mM sodium fluoride as a phosphatase inhibitor, 1× protease inhibitor cocktail (add before each cell lysis).
-
2.
W6/32 antibody: antibody purified from the conditioned media of HB-95 mouse hybridoma that recognizes human MHC I (HLA-A, B, C).
-
3.
Cyanogen bromide (CNBr)-activated agarose or sepharose beads.
-
4.
Phosphate buffered saline (PBS): purchased pre-prepared.
-
5.
100 mM Tris(hydroxymethyl)aminomethane hydrochloride (100 mM Tris–HCl).
-
6.
10% (v/v) acetic acid.
2.3 MHC Peptide Preconcentration and Desalting
For MHC peptide purification, LC-MS grade water is needed to prepare all solutions:
-
1.
Acidifying solution: 10% (v/v) trifluoroacetic acid (TFA).
-
2.
Elution buffer: 30% (v/v) acetonitrile with 0.1% (v/v) TFA.
-
3.
Washing buffer: 0.1% (v/v) TFA.
-
4.
C18 solid phase extraction (SPE) cartridge.
2.4 Enrichment of Phosphorylated MHC Peptides
All reagents must be analytical or higher grade unless specified.
-
1.
MHC immunopeptidome isolated from A549 cells.
-
2.
Nitrilotriacetic acid (NTA) silica with a particle size of 16–24 μm.
-
3.
Iron (III) chloride (FeCl3).
-
4.
Titanium dioxide nanoparticles, 20 nm in particle size.
-
5.
C18 spin column.
-
6.
Acetonitrile (ACN), LC-MS grade.
-
7.
Water, LC-MS grade.
-
8.
Dihydroxybenzoic acid (DHB).
-
9.
Formic acid (FA), LC-MS grade.
-
10.
Ethylenediaminetetraacetic acid (EDTA), analytical grade.
-
11.
Trifluoroacetic acid (TFA), analytical grade.
-
12.
25% ammonia solution in water.
-
13.
Acetic acid, analytical grade.
-
14.
IMAC resin washing buffer 1: 0.01% (v/v/) acetic acid.
-
15.
IMAC resin washing buffer 2: 0.01% (v/v) acetic acid, 15 ACN.
-
16.
Fe3+ IMAC loading buffer: 0.1 M acetic acid.
-
17.
Fe3+ IMAC washing buffer: 30% (v/v) ACN, 0.07% (v/v) TFA.
-
18.
Fe3+ IMAC eluting buffer: 0.5% (v/v) ammonia.
-
19.
TiO2 loading buffer: 80% (v/v) ACN, 6% (v/v) TFA and 50 mg/mL DHB.
-
20.
TiO2 washing buffer: 50% (v/v) ACN, 0.1% (v/v) TFA.
-
21.
TiO2 eluting buffer: 5% (v/v) ammonia.
2.5 LC-MS/MS Analysis
Q Exactive mass spectrometer coupled with a nano-LC is recommended for the analysis of MHC peptides. The nano-LC column can be packed in-house with 1.9 μm reverse phase beads (75 μm i.d., length ≥ 20 cm) or columns that are commercially available, e.g., Acquity BEH C18 column from Waters, Acclaim PepMap 100 C18 LC columns from Thermo, etc.
2.6 Database Search
-
1.
Standard Protein database: UniProt human protein database.
-
2.
Customized database: In-house-built protein database constructed using exome sequencing data [20].
-
3.
Database search engine: Mascot (Matrix Science, version 2.3 or later).
3 Methods
3.1 Cell Culture
Grow A549 cells in cell culture media at 37 °C with 5% CO2 in T175 flask until 80% confluent. Collect cells from ten flasks for one MHC peptide isolation experiment (see Note 1 ).
3.2 Immuno-precipitation
-
1.
Couple W6/32 antibody to CNBr-activated agarose or sepharose by incubation of antibody and beads overnight at 4 °C. Overnight incubation must be with rotation. Rinse the beads, and block the unreacted CNBr group with 100 mM Tris–HCl buffer three times. The prepared beads can be stored in PBS at 4 °C.
-
2.
Add 10 mL of cold PBS to harvested cells. Centrifuge at 600 × g for 3 min wash and discard supernatant. Repeat a total of three times.
-
3.
Add 2 mL of cell lysis buffer with fresh protease inhibitor and phosphatase inhibitor to each flask, and detach the cells by gently shaking the flask.
-
4.
Transfer the detached cells in lysis buffer to a glass homogenizer. Break the cells with loose piston, and transfer the homogenate to a 50 mL tube (see Note 2 ).
-
5.
Centrifuge the homogenate at 20,000 × g for 15 min and keep the supernatant. Store 100 μL of lysate for western blotting (see Note 3 ), and use the rest for immunoprecipitation.
-
6.
Incubate the lysate with 1 mL of agarose beads (W6/32 antibody conjugated) overnight at 4 °C with gentle rotation.
-
7.
Centrifuge at 600 × g for 3 min to pellet the beads. Discard the supernatant and wash the beads with the same volume of washing buffer as that used for cell lysis. Repeat wash three times.
-
8.
Elute the bound MHC by adding 5 mL of 10% acetic acid to the agarose beads, and incubate at room temperature for 15 min. Centrifuge at 600 × g for 3 min and collect the supernatant as elution. If desired, retain 100 μL of the elution for western blotting confirmation of MHC isolation, and store the rest in −80 °C until LC-MS analysis (see Note 3 ).
3.3 MHC Peptide Preconcentration and Desalting
-
1.
Add acidifying solution to sample solution to adjust TFA concentration at 0.1% (i.e., 50 μL of acidifying solution to 5 mL of elution).
-
2.
Activate the C18 cartridge with 1 mL of ACN.
-
3.
Clean the C18 cartridge with 1 mL of washing buffer.
-
4.
Load acidified sample solution from step 1 to the C18 cartridge.
-
5.
Wash the C18 cartridge with 1 mL washing buffer three times.
-
6.
Elute the bound peptides with 1 mL of elution buffer.
-
7.
Evaporate eluted peptides to dryness using a Speed Vac.
-
8.
Dissolve the dried peptides in 1 mL washing buffer and repeat the steps from 2 to 7. The MHC peptides are ready for phosphopeptide enrichment.
3.4 Preparation of Iron (III)-NTA IMAC
-
1.
Reconstitute the NTA silica with water (1 mg/mL) in a 1.5 mL Eppendorf tube.
-
2.
Centrifuge at 13,000 × g for 5 min, and discard the supernatant.
-
3.
Add 1 mL of 50 mM ETDA and vortex for 5 min at room temperature.
-
4.
Centrifuge at 13,000 × g for 5 min, discard the supernatant.
-
5.
Repeat step 2 twice.
-
6.
Activate the resin by adding 1 mL of 100 mM FeCl3 and stirring overnight.
-
7.
Centrifuge at 13,000 × g for 5 min, discard the supernatant.
-
8.
Wash the resin with 1 mL IMAC resin washing buffer 1, followed by wash with IMAC resin washing buffer 2 and finally with another wash with IMAC resin washing buffer 1.
-
9.
Store the Fe-NTA IMAC resin in 0.01% acetic acid at 4 °C at the concentration of 50 mg/mL. The resin can be stored for up to 1 month.
3.5 Enrichment of Phosphorylated MHC Peptides with IMAC
-
1.
Reconstitute immune-purified HLA-associated peptide samples in 100 μL Fe3+ IMAC loading buffer.
-
2.
Add 20 μL of Fe3+ IMAC suspension to the MHC peptide sample solution, and vortex for 30 min (see Note 4 ).
-
3.
Centrifuge at 13,000 × g for 5 min, discard the supernatant.
-
4.
Add 500 μL of Fe3+ IMAC washing buffer and vortex for 15 min (see Note 5 ).
-
5.
Centrifuge at 13,000 × g for 5 min, discard the supernatant.
-
6.
Adding 100 μL of Fe3+ IMAC elution buffer and vortex for 15 min.
-
7.
Centrifuge at 13,000 × g for 5 min, transfer the supernatant to a clean tube.
-
8.
Add 100 μL of acidifying solution.
-
9.
Activate C18 spin column by adding 50 μL of ACN, spin at 1000 × g for 1 min.
-
10.
Wash the spin column with 50 μL of washing buffer.
-
11.
Load the peptides onto the spin column, spin at 1000 × g for 1 min.
-
12.
Wash the spin column with 50 μL of washing buffer.
-
13.
Elute desalted peptides with 50 μL elution buffer.
-
14.
Dry the peptides eluted from the C18 column using a Speed Vac. Store the dried samples at −80 °C until LC-MS analysis.
3.6 Enrichment of Phosphorylated MHC Peptides with TiO2 Nanoparticles
-
1.
Preparation of TiO2 nanoparticles suspension of 20 mg/mL in 6% TFA, 80% acetonitrile (see Notes 6 and 7 ).
-
2.
Reconstitute purified MHC peptides in 200 μL of loading buffer.
-
3.
Adding 10 μL of TiO2 suspension to the MHC peptides solution.
-
4.
Shake at room temperature for 30 min.
-
5.
Centrifuge at 10,000 × g for 5 min, discard supernatant.
-
6.
Add 500 μL of washing buffer and vortex for 15 min.
-
7.
Centrifuge at 10,000 × g for 5 min, discard supernatant.
-
8.
Add 200 μL of 5% ammonia, shake for 15 min.
-
9.
Centrifuge at 10,000 × g for 5 min, transfer to supernatant to a clean tube without disturbing the pellet.
-
10.
Acidify eluted peptides solution with 100 μL of acidifying solution.
-
11.
Desalt the eluted peptides with C18 spin column as described in Subheading 3.5.
-
12.
Dry the eluted peptides using a Speed Vac, and store the samples at −80 °C until MS analysis.
3.7 LC-MS/MS Analyses
-
1.
Nano-LC separation: A 2-h gradient was performed from 5% to 35% acetonitrile (v/v) containing 0.1% formic acid (v/v) at a flow rate between 200 nL/min and 500 nL/min.
-
2.
Data-dependent MS/MS acquisition: The data acquisition method consisted of one full MS scan from m/z 300 to 1500, followed by data-dependent MS/MS scan for up to 12 most intense ions with a dynamic exclusion (repeat count of two and repeat exclusion duration of 30 s). Charge selection was enabled to select +2 and +3 charged precursors for MS/MS. All data were recorded with the Xcalibur software and exported as .raw files for further analysis.
3.8 Database Search and Data Processing
-
1.
Raw files were transformed to .mgf files with ProteoWizard [21].
-
2.
Search with Mascot (Matrix Science Inc.) against the latest UniProt human database with reversed sequence as decoy database. Mass tolerance was set as 8 ppm for precursor and 0.05 Da for fragment ions. No enzyme digest was selected.
-
3.
Set methionine oxidation (+15.994915) and phosphorylation at serine, threonine, and tyrosine (+79.966331) as variable modification.
-
4.
After database searching, peptides-spectra matches (PSM) were filtered with the following criteria: top 1 match, Mascot Score ≥ 22, and 8–14 amino acids in length. False discovery rate (FDR) was calculated as the following: FDR = [(PSM)decoy × 2]/[(PSM)target + (PSM)decoy]. An FDR value between 5 and 9% would be expected (see Notes 8 and 9 ).
4 Notes
-
1.
The amount of cells used for phosphopeptide enrichment can be increased up to 1 × 109, to ensure sufficient starting material for MS detection of phosphopeptides. For the optimization of the phosphopeptide enrichment procedure, the starting amount could be lower. However, when working with tissue samples, the amount of starting material is sample-dependent as the expression of HLA from each sample has large variation.
-
2.
Phosphatase inhibitors are added to the lysis buffer for phosphopeptide enrichment to prevent dephosphorylation of peptides. Phosphatase inhibitor cocktail is also available from a number of commercial sources.
-
3.
We suggest retaining 100 μL of the elution for SDS-PAGE and western blotting to confirm success of immuno pulldown of MHCs. We suggest running an SDS-PAGE of the original cell lysate, flow-through, wash, and elution from immunoprecipitation. After standard western blot transfer to membranes, we typically probe with 1:2000 diluted antihuman HLA antibody overnight at 4 °C with constant rotation and then 1:5000 diluted secondary antibody for detection. The intensity of blots from flow-through, wash, and elution is compared to verify the efficiency of immunoprecipitation.
-
4.
Off-line SPE was used in this protocol. However, it has been reported that the specificity of enrichment could be increased by packing the material in a spin column or using a high-performance liquid chromatograph (HPLC). We chose off-line SPE as it is easy to operate and does not require specialized instrumentation.
-
5.
For both IMAC and MOAC, the volume of washing buffer and number of washing steps might need to be optimized for each sample. This is dependent upon the complexity of the samples. Extra washing is needed if the specificity of phosphopeptide identification is low. Since immunopeptidome is less complex than tryptic proteome, only one wash is used in this protocol to minimize sample loss.
-
6.
TiO2 with larger particles sizes (up to 50 μm) can be used for phosphopeptide enrichment. Other metal oxide, e.g., ZrO2, may also provide complementary coverage.
-
7.
Lactic acid and glycolic acid are alternatives for DHB in the loading buffer to reduce the non-specific binding of peptides containing acidic amino acids.
-
8.
The presence of phosphate group can be verified by the neutral loss in MS/MS spectrum, but the intensity is dependent on peptide sequence.
-
9.
A lower FDR setting (e.g., 5%) can be used as initial PSM filter, and extra criteria (e.g., peptide length, binding affinity to HLA) need to remove possible false identifications. For lower PSM scores, we suggest manual inspection of the MS/MS spectra to confirm amino acid sequences.
References
Hunter T (2000) Signaling—2000 and beyond. Cell 100:113–127
Ubersax JA, Ferrell JE Jr (2007) Mechanisms of specificity in protein phosphorylation. Nat Rev Mol Cell Bio 8:530–541
Brognard J, Hunter T (2011) Protein kinase signaling networks in cancer. Curr Opin Genet Dev 21:4–11
Huttlin EL, Jedrychowski MP, Elias JE, Goswami T, Rad R, Beausoleil SA et al (2010) A tissue-specific atlas of mouse protein phosphorylation and expression. Cell 143:1174–1189
Francavilla C, Lupia M, Tsafou K, Villa A, Kowalczyk K, Jersie-Christensen RR et al (2017) Phosphoproteomics of primary cells reveals druggable kinase signatures in ovarian cancer. Cell Rep 18:3242–3256
Mohammed F, Cobbold M, Zarling AL, Salim M, Barrett-Wilt GA, Shabanowitz J et al (2008) Phosphorylation-dependent interaction between antigenic peptides and MHC class I: a molecular basis for the presentation of transformed self. Nat Immunol 9:1236–1243
Locard-Paulet M, Lim L, Veluscek G, McMahon K, Sinclair J, Van Weverwijk A et al (2016) Phosphoproteomic analysis of interacting tumor and endothelial cells identifies regulatory mechanisms of transendothelial migration. Sci Signal 9:ra15-ra15
Yi T, Zhai B, Yu Y, Kiyotsugu Y, Raschle T, Etzkorn M et al (2014) Quantitative phosphoproteomic analysis reveals system-wide signaling pathways downstream of SDF-1/CXCR4 in breast cancer stem cells. Proc Natl Acad Sci U S A 111:E2182–E2190
K-L H, Li S, Mertins P, Cao S, Gunawardena HP, Ruggles KV et al (2017) Proteogenomic integration reveals therapeutic targets in breast cancer xenografts. Nat Commun 8:14864
Dazert E, Colombi M, Boldanova T, Moes S, Adametz D, Quagliata L et al (2016) Quantitative proteomics and phosphoproteomics on serial tumor biopsies from a sorafenib-treated HCC patient. Proc Natl Acad Sci U S A 113:1381–1386
Zarling AL, Polefrone JM, Evans AM, Mikesh LM, Shabanowitz J, Lewis ST et al (2016) Identification of class I MHC-associated phosphopeptides as targets for cancer immunotherapy. Proc Natl Acad Sci U S A 103:14889–14894
Bassani-Sternberg M, Bräunlein E, Klar R, Engleitner T, Sinitcyn P, Audehm S et al (2016) Direct identification of clinically relevant neoepitopes presented on native human melanoma tissue by mass spectrometry. Nat Commun 7:13404
Laumont CM, Daouda T, Laverdure J-P, Bonneil É, Caron-Lizotte O, Hardy M-P et al (2016) Global proteogenomic analysis of human MHC class I-associated peptides derived from non-canonical reading frames. Nat Commun 7:10238
Olsen JV, Mann M (2013) Status of large-scale analysis of post-translational modifications by mass spectrometry. Mol Cell Proteomics 12:3444–3452
Andersson L, Porath J (1986) Isolation of phosphoproteins by immobilized metal (Fe3+) affinity chromatography. Anal Biochem 154:250–254
Pinkse MW, Uitto PM, Hilhorst MJ, Ooms B, Heck AJ (2004) Selective isolation at the femtomole level of phosphopeptides from proteolytic digests using 2D-NanoLC-ESI-MS/MS and titanium oxide precolumns. Anal Chem 76:3935–3943
Zhou H, Low TY, Hennrich ML, van der Toorn H, Schwend T, Zou H et al (2011) Enhancing the identification of phosphopeptides from putative basophilic kinase substrates using Ti (IV) based IMAC enrichment. Mol Cell Proteomics 10:M110.006452
Potel CM, Lin M-H, Heck AJ, Lemeer S (2018) Defeating major contaminants in Fe3+-IMAC phosphopeptide enrichment. Mol Cell Proteomics 17:1028–1034
Abelin JG, Trantham PD, Penny SA, Patterson AM, Ward ST, Hildebrand WH et al (2015) Complementary IMAC enrichment methods for HLA-associated phosphopeptide identification by mass spectrometry. Nat Protoc 10:1308–1318
Chen R, Fauteux F, Foote S, Stupak J, Tremblay T-L, Gurnani K et al (2018) Chemical derivatization strategy for extending the identification of MHC class I immunopeptides. Anal Chem 90:11409–11416
Chambers MC, Maclean B, Burke R, Amodei D, Ruderman DL, Neumann S et al (2012) A cross-platform toolkit for mass spectrometry and proteomics. Nat Biotechnol 30:918–920
Author information
Authors and Affiliations
Corresponding author
Editor information
Editors and Affiliations
Rights and permissions
Copyright information
© 2019 Springer Science+Business Media, LLC, part of Springer Nature
About this protocol
Cite this protocol
Chen, R., Li, J. (2019). Enrichment of Phosphorylated MHC Peptides with Immobilized Metal Affinity Chromatography and Titanium Dioxide Particles. In: Fulton, K., Twine, S. (eds) Immunoproteomics. Methods in Molecular Biology, vol 2024. Humana, New York, NY. https://doi.org/10.1007/978-1-4939-9597-4_16
Download citation
DOI: https://doi.org/10.1007/978-1-4939-9597-4_16
Published:
Publisher Name: Humana, New York, NY
Print ISBN: 978-1-4939-9596-7
Online ISBN: 978-1-4939-9597-4
eBook Packages: Springer Protocols