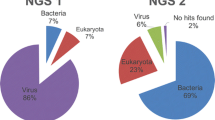
Abstract
While classical virology techniques such as virus culture, electron microscopy, or classical PCR had been unsuccessful in identifying the causative agent responsible for the fulminating disease of guinea fowl, we identified a novel avian gammacoronavirus associated with the disease using metagenomics. Next-generation sequencing is an unbiased approach that allows the sequencing of virtually all the genetic material present in a given sample.
You have full access to this open access chapter, Download protocol PDF
Similar content being viewed by others
Key words
1 Introduction
The field of pathogen discovery gained a completely new dimension when new genomics tools, next-generation sequencing (NGS), became available. Isolation and identification by electronic microscopy used to be the gold standard techniques to identify a new pathogen. However, some pathogens are difficult to culture, and/or to separate from co-infecting agents. PCR has improved pathogen discovery, but even when pan-species/Genus/family PCRs may be developed (which is not always possible, especially for highly variable RNA viruses), the primers chosen do condition the nucleic acids that will be amplified: it is still a “biased” technique with which only known (or closely related to known) pathogens whose presence was suspected may be detected [1, 2]. NGS allows for (1) massive sequencing of genetic material and (2) unbiased sequencing, to a much lower cost per sequenced base than Sanger sequencing technology. Here we describe an NGS technique that can be used to identify novel pathogens, which we recently used to identify the pathogen responsible for the guinea fowl fulminating disease [3].
2 Materials
2.1 Materials for Preparation of Specimens and Concentrating Viral Particles
-
1.
PBS.
-
2.
PBS containing 100 U/ml Penicillin and 0.1 mg/ml streptomycin.
-
3.
0.45 μm filter.
-
4.
RNAse.
-
5.
DNAse (10 U/μl).
-
6.
10× DNase buffer: 200 mM Tris–HCl (pH 8.4), 20 mM MgCl2, 500 mM KCl.
2.2 Materials for RNA Extraction and Amplification
-
1.
TRIzol reagent or similar.
-
2.
Chloroform.
-
3.
RNA extraction kit, e.g., Nucleospin RNA virus kit (Macherey-Nagel) or similar.
-
4.
Reverse transcription kit, e.g., RevertAid kit (Thermofisher) or similar.
-
5.
10 mM dNTPs.
-
6.
RNase inhibitor.
-
7.
20 μM tagged random hexamer: 454-A: 5′ ATG-GTC-GTC-GTA-GGC-TGC-TCN-NNN-NNN-N 3′ (see Note 1 ).
-
8.
PCR kit, e.g., Phusion (NEB) or similar.
-
9.
20 μM PCR primer (tag only): 454-A: 5′ ATG-GTC-GTC-GTA-GGC-TGC-TC 3′.
-
10.
Thermocycler.
-
11.
1 % agarose gel.
-
12.
1× TBE buffer: 89 mM Tris, 89 mM orthoboric acid, 2 mM EDTA.
-
13.
Sybr® safe (Life technologies) or similar.
-
14.
1 kb plus DNA ladder.
-
15.
Gel purification kit.
-
16.
PicoGreen® quantitation assay (Life technologies) or similar.
2.3 Next-Generation Sequencing
-
1.
Miseq reagent kit (Illumina).
-
2.
Miseq (Illumina).
3 Methods
Figure 1 summarizes the methodology used for the identification of a novel guinea fowl gammacoronavirus.
3.1 Sample Preparation
-
1.
Pool intestinal contents of experimentally infected guinea fowl poults and resuspend in 500 μl PBS with penicillin and streptomycin. Vortex.
-
2.
Filter (0.45 μm filter) the solution to eliminate eukaryotic- and bacterial-cell-sized particles.
-
3.
Centrifuge the digestive content at 10,000 × g for 30 min twice to clarify the solution and collect the supernatant in a new tube.
3.2 Concentration of Viral Particles
-
1.
Pellet the concentrated material by ultracentrifugation at 100,000 × g for 2 h.
-
2.
Treat with RNAse and DNAse to remove non-particle-protected nucleic acids: make a mix of 500 μl of sample, 10 μl DNAse (100 U), 12 μl RNAse (20 μg/μl), 60 μl 10× DNAse buffer, 16 μl PBS. Incubate the mix for 20 min at 37 °C and then 10 min at 75 °C to stop the reaction.
3.3 RNA Extraction and Amplification (See Note 2 )
-
1.
Add 750 μl TRIzol to 250 μl sample from Subheading 3.2, step 2 and incubate 5 min at room temperature.
-
2.
Add 200 μl chloroform, vortex vigorously, and incubate 10 min at room temperature.
-
3.
Centrifuge for 15 min at 11,000 × g at 4 °C.
-
4.
Collect the top aqueous phase.
-
5.
Extract RNA from 150 μl of the collected aqueous phase on a silicate column with a RNA extraction kit.
-
6.
Perform a reverse transcription reaction using random primers by mixing 7.5 μl RNA, 5 μl tagged random hexamer.
-
7.
Incubate at 65 °C for 5 min then keep on ice.
-
8.
Add 4 μl 5× reaction buffer, 0.5 μl RNase inhibitor, 2 μl dNTP, and 1 μl RevertAid reverse transcriptase.
-
9.
Incubate at 25 °C for 10 min, 42 °C for 60 min, and 70 °C for 10 min.
-
10.
Reactions can now be stored at −20 °C or used immediately for PCR.
-
11.
Perform a random PCR by mixing 10 μl 5× Phusion HF reaction buffer, 1 μl dNTP, 0.5 μl tag only primer, 5 μl cDNA, 0.5 μl Phusion polymerase, and 33 μl water.
-
12.
Perform PCR using the following cycle: 98 °C 30 s followed by 40 cycles of 98 °C 10 s, 65 °C 20 s, and 72 °C 30 s followed by a final incubate of 72 °C 10 min.
-
13.
Analyze the PCR products on a 1 % agarose gel, migrate for 1 h at 60 V.
-
14.
Excise the 300 bp bands and perform a gel purification with a commercial kit.
-
15.
Quality assessment of the prepared library: quantify the DNA generated by a fluorescence-based method (PicoGreen® quantitation assay) and aim at 1 μg DNA as input; check the DNA quality and aim for a 260:280 ratio >1.8.
3.4 Cluster Generation Using the Miseq Reagent Kit (Illumina)
-
1.
Hybridize sample to flow cell.
-
2.
Amplify sample (bridge amplification).
-
3.
Linearize fragments.
-
4.
Block fragments.
-
5.
Hybridize sequencing primer.
3.5 Sequencing on the Miseq (Illumina)
-
1.
The library DNA fragment act as a template, from which a complementary strand is synthesized.
-
2.
ddNTPs are added one by one (one cycle = one ddNTP added, a picture taken and defluoration of the ddNTP to be able to add a new ddNTP the next cycle) by a DNA polymerase. The addition of ddNTP is digitally recorded as sequence data cycle after cycle.
3.6 Data Analysis
-
1.
Preprocess the data to remove adapter sequences and demultiplexing using splitbc (several samples can be multiplexed and run together on the MiSeq Illumina sequencer to reduce cost).
-
2.
Preprocess the data to remove low-quality reads and compiling paired sequences using illuminapairedend.
-
3.
Map the data to a reference genome or de novo align the sequence reads (alignment with bwa [4], consensus computed with the SAMtools [5] software package. Display the results with the IGV [6] browser).
-
4.
Analyze the compiled sequence with the GAAS software (http://gaas.sourceforge.net/) with an expected value of 10−3.
4 Notes
-
1.
Victoria et al. [7] described a panel of tagged primers (named with alphabet letter), and while we selected “454-A” for the present study, any of the tagged primers described in [7] could be used instead (454-B, 454-C, etc).
-
2.
To identify pathogens with DNA genomes, a DNA extraction would be performed, followed by a Klenow step before the random PCR:
-
(a)
DNA extraction: High Pure template preparation kit (Roche) can be used, following the manufacturer’s instructions.
-
(b)
Klenow step on DNA: mix 2.5 μl tagged random primer, 3 μl 10× buffer, 1 μl 0.5 mM dNTP, 10 μl DNA, 1 μl 0.5 U/μl DNA pol1 and 12.5 μl water. Incubate at room temperature for 1 h.
-
(c)
Proceed from Subheading 3.3, step 10.
-
(a)
References
Padmanabhan R, Mishra AK, Raoult D et al (2013) Genomics and metagenomics in medical microbiology. J Microbiol Methods 95:415–424
Miller RR, Montoya V, Gardy JL et al (2013) Metagenomics for pathogen detection in public health. Genome Med 5:81
Liais E, Croville G, Mariette J et al (2014) Novel avian coronavirus and fulminating disease in guinea fowl, France. Emerg Infect Dis 20:105–108
Li H, Durbin R (2009) Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics 25:1754–1760
Li H, Handsaker B, Wysoker A et al (2009) The Sequence alignment/map format and SAMtools. Bioinformatics 25:2078–2079
Robinson JT, Thorvaldsdóttir H, Winckler W et al (2011) Integrative genomics viewer. Nat Biotechnol 29:24–26
Victoria JG, Kapoor A, Li L et al (2009) Metagenomic analyses of viruses in stool samples from children with acute flaccid paralysis. J Virol 83:4642–4651
Author information
Authors and Affiliations
Corresponding author
Editor information
Editors and Affiliations
Rights and permissions
Copyright information
© 2015 Springer Science+Business Media New York
About this protocol
Cite this protocol
Ducatez, M.F., Guérin, JL. (2015). Identification of a Novel Coronavirus from Guinea Fowl Using Metagenomics. In: Maier, H., Bickerton, E., Britton, P. (eds) Coronaviruses. Methods in Molecular Biology, vol 1282. Humana Press, New York, NY. https://doi.org/10.1007/978-1-4939-2438-7_2
Download citation
DOI: https://doi.org/10.1007/978-1-4939-2438-7_2
Published:
Publisher Name: Humana Press, New York, NY
Print ISBN: 978-1-4939-2437-0
Online ISBN: 978-1-4939-2438-7
eBook Packages: Springer Protocols