Abstract
Mammalian cell nuclei contain multiple granular structures, which are termed nuclear bodies. These structures are involved in various molecular events in the nucleus; they provide platforms for biogenesis of macromolecular complexes that are essential for gene expression, such as the ribosome and spliceosome; they act as reservoirs of various regulatory factors; and they are involved in the regulation of specific gene loci. Nuclear bodies are usually visualized by immunostaining for specific marker proteins. Although each type of nuclear body contains a distinct set of proteins, the protein components of most types of nuclear bodies remain to be identified. This chapter introduces a new approach to identify the protein components of specific types of nuclear bodies.
Similar content being viewed by others
Key words
1 Introduction
The nuclei of mammalian cells are highly organized. Each nucleus contains nuclear bodies, which are membraneless organelles that contain specific proteins or RNAs characteristic of particular nuclear processes [1, 2]. Nuclear bodies provide a platform for the biogenesis of fundamental macromolecular machineries. For example, the nucleolus and the Cajal body provide a platform for the ribosome and spliceosome, respectively [3, 4]. Interchromatin granules (ICGs) (also known as speckles) contain various splicing factors, such as the members of the serine/arginine-rich (SR) protein family. ICGs act as reservoirs of these factors, enabling them to be recruited to specific chromosomal loci upon transcriptional activation [5]. It was recently reported that growth control genes relocate between ICGs and Polycomb bodies in response to growth signals [6]. Other types of nuclear bodies participate in the regulation of specific genes, e.g., the histone locus body forms around histone gene clusters, at which regulation of histone gene transcription and subsequent processing of histone mRNAs occur in a cell-cycle-dependent manner [4]. PML bodies are implicated in DNA repair because they recruit and release many of the proteins that are involved in this process [7].
Nuclear bodies are usually visualized by immunostaining for specific marker proteins (Table 1). Several relatively abundant, long noncoding RNAs (lncRNAs) were recently found to localize to specific types of nuclear bodies [8–16]. Therefore, these types of nuclear bodies can also be visualized by in situ hybridization of lncRNAs. Among these lncRNAs, nuclear paraspeckle assembly transcript 1 localizes specifically to paraspeckles and is an essential structural component of these nuclear bodies [11–14]. Therefore, paraspeckles can be considered to be huge ribonucleoprotein complexes. Other lncRNAs have been identified that are required for the formation of other types of nuclear bodies [9, 16]. Multiple proteins interact with these lncRNAs, and these proteins are thereby sequestrated in nuclear bodies, which is critical to control the functional activities of the proteins.
It is important to identify the components of nuclear bodies to understand their biological functions and the mechanisms underlying their actions. To identify the components of a distinct type of nuclear body, the most straightforward approach is to purify these nuclear bodies and to characterize their components using proteomics analysis. Indeed, protocols to purify nucleoli and ICGs have been established, and the protein components of these structures have been extensively characterized [17, 18]. However, some types of nuclear bodies appear to be unstable, meaning they cannot be easily purified because their structure does not remain intact during the purification procedure. Some types of nuclear bodies fractionate into an insoluble nuclear matrix fraction that is usually difficult to solubilize without disrupting their structures. Moreover, there is no technique to assess whether each type of nuclear body remains structurally intact following purification.
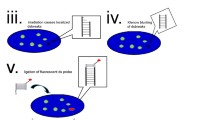
Here, we describe a novel method called “colocalization screening” to identify novel protein components of distinct types of nuclear bodies. This method uses human protein expression (HUPEX) clones constructed from the original human full-length (FLJ) cDNA clones [19], each of which expresses a protein that is fused to Venus, a fluorescent protein tag [21]. Venus-tagged proteins that colocalize with immunostaining for an endogenous marker of a specific type of nuclear body (Table 1) are selected as candidate nuclear body components (Fig. 1). Prior to experimental screening, candidate clones that encode Venus-tagged proteins that localize to nuclear foci should be selected from the Human Gene and Protein Database (HGPD) (see Note 1 ) [20, 21]. The HGPD contains the entire dataset concerning the intracellular localizations of 32651 Venus-tagged FLJ proteins (see Note 2 ). Using this method, we identified 35 novel components of paraspeckles (Fig. 2) [22]. In total, 68 Venus-tagged proteins localized to paraspeckle-like nuclear foci. Immunostaining with an antibody against the paraspeckle marker protein splicing factor proline/glutamine rich (SFPQ) was performed. Each Venus-tagged protein was confirmed to label paraspeckles if its signal colocalized with that of this endogenous marker protein (Fig. 2). This colocalization screening method is a powerful tool that can be used to identify the protein components of other types of nuclear bodies by performing immunolabeling for various endogenous markers. We describe how to perform this colocalization screening procedure using HeLa cells to identify novel nuclear body proteins, although the procedure is also applicable to other cell lines.
Outline of the colocalization screening of nuclear body components. Following transfection of a full-length (FLJ) cDNA clone, cells will overexpress a candidate nuclear body protein. Venus signals are prominent in some nuclear foci (green foci in the center of the cell). Colocalization screening to select FLJ clones encoding Venus-tagged proteins that colocalize with an endogenous marker protein of nuclear foci, which is labeled by immunostaining (red foci in the bottom of the cell). The red arrow indicates colocalization of the Venus signal with that of the endogenous marker
Images of colocalization screening to identify novel paraspeckle proteins. Venus signals (green) from two FLJ clones (FLJ22617 and FLJ27264) colocalize with the immunostaining signals of endogenous SFPQ (magenta), a paraspeckle marker protein. Therefore, Ewing sarcoma breakpoint region 1 (EWSR1) and fused in sarcoma (FUS), which are expressed by FLJ22617 and FLJ27264, respectively, were selected as candidate paraspeckle proteins. The localization of endogenous EWSR and FUS to paraspeckles was confirmed by immunostaining using antibodies against these proteins [22]. An image in which a Venus-tagged protein (COIL-Venus) does not colocalize with endogenous SFPQ, but localizes to another type of nuclear body, is shown on the right as a reference. Bar, 0.5 μm
2 Materials
All solutions were prepared using ultrapure water (prepared by purifying deionized water to attain a conductivity of 18.2 MΩ cm at 25 °C) and analytical grade reagents.
2.1 Plasmid Preparation
-
1.
Escherichia coli stocks harboring FLJ clone plasmids (Venus-tagged expression clones) (see Notes 3 and 4 ).
-
2.
Luria broth (LB) medium (Nacalai Tesque).
-
3.
Ampicillin stock solution (50 mg/mL).
-
4.
QIAGEN Plasmid Midi Kit (QIAGEN).
2.2 Cell Culture
-
1.
HeLa (human cervical cancer) cells (see Note 5 ).
-
2.
Dulbecco’s Modified Eagle Medium (DMEM) (see Note 6 ).
-
3.
Fetal bovine serum (FBS).
-
4.
Trypsin-EDTA.
-
5.
Hemocytometer (SLGC).
-
6.
8-well chamber slide (e.g., Lab-TekII Chamber Slide, Thermo Fisher Scientific).
2.3 Transfection
-
1.
FLJ plasmids.
-
2.
Lipofectamine 2000 (Invitrogen, Life Technologies).
-
3.
Opti-MEM I Reduced-Serum Medium (Gibco, Invitrogen Life, Technologies).
2.4 Immunofluorescence
-
1.
Phosphate buffered saline (PBS) [137 mM NaCl, 2.7 mM KCl, 10 mM Na2HPO4 2H2O, KH2PO4 (pH 7.4)].
-
2.
PBS containing 4 % paraformaldehyde (see Note 7 ).
-
3.
TBS [50 mM Tris-Cl, pH 7.5, 150 mM NaCl].
-
4.
TBS-Triton X-100 [50 mM Tris-Cl, pH 7.5, 150 mM NaCl, 0.2 % Triton X-100].
-
5.
TBST [50 mM Tris-Cl, pH 7.5, 150 mM NaCl, 0.05 % Tween 20].
-
6.
TBS containing 10 % normal horse serum (VECTOR Laboratories Inc.).
-
7.
Primary antibody (see Note 8 ).
-
8.
Secondary antibody (conjugated to Alexa Fluor 568) (see Note 9 ).
-
9.
VECTASHIELD Mounting Medium with DAPI (VECTOR Laboratories, Inc.).
3 Methods
3.1 Data Mining of the HGPD
-
1.
Obtain information about FLJ clones that express proteins that potentially localize to particular types of nuclear foci (see Note 10 ).
-
2.
Access the HGPD (http://hgpd.lifesciencedb.jp/) to collect images of the localizations of the proteins expressed by the selected FLJ clones (see Note 11 ).
-
3.
If possible, narrow down the FLJ clones that are to be used for colocalization screening by examining the features of the nuclear foci in these images (Fig. 1) (see Note 12 ).
3.2 Plasmid Preparation
-
1.
Streak bacterial stocks of FLJ clones onto a LB culture dish and incubate overnight at 37°C.
-
2.
Pick a single isolated colony and incubate in 2 mL of LB medium containing 50 μg/mL ampicillin at 37 °C for ~12 h.
-
3.
Transfer the starter culture to 50 mL of LB medium containing 50 μg/mL ampicillin, and culture at 37 °C for 16 h.
-
4.
Purify plasmids using the QIAGEN Plasmid Midi Kit according to the instruction manual.
-
5.
Determine the plasmid concentration by measuring absorbance at 260 nm (A 260) using a spectrophotometer.
-
6.
Adjust the plasmid concentration to 37.5 ng/μL with distilled water.
3.3 Cell Culture
-
1.
Thaw frozen cell stocks in a water bath at 37 °C. Centrifuge stocks at a low speed (220 × g, 5 min) to pellet cells. Remove the supernatant by pipetting.
-
2.
Gently resuspend the cell pellet in a growth medium (DMEM supplemented with 10 % FBS).
-
3.
Culture cells in a growth medium in a humidified incubator with 5 % CO2 until they reach 80 % confluency.
3.4 Plasmid Transfection
-
1.
Seed 2 × 104 cells in 700 μL of growth medium into each well of an 8-well chamber slide 1 day prior to transfection. Cells should be transfected when they have reached 30–50 % confluency.
-
2.
Add 2 μL of Venus-tagged FLJ clone plasmid (75 ng) to 40 μL Opti-MEM I Reduced-Serum Medium.
-
3.
In a separate tube, add 0.25 μL of Lipofectamine 2000 reagent to 40 μL Opti-MEM I Reduced-Serum Medium.
-
4.
Combine the solutions prepared in steps 2 and 3, and incubate the mixture for 10–20 min at room temperature to form plasmid-Lipofectamine 2000 complexes.
-
5.
Add the resultant complexes (ca. 80 μL) directly to the growth medium in the chamber slide (see Note 13 ).
-
6.
Culture the cells for 17 h in a humidified incubator with 5 % CO2 (see Note 14 ).
3.5 Immunofluorescence
-
1.
Remove the culture medium by aspiration and wash the cells once with PBS.
-
2.
Fix the cells with PBS containing 4 % paraformaldehyde for 10 min at room temperature.
-
3.
Permeabilize the fixed cells with TBS-Triton X-100 for 5 min.
-
4.
Rinse the samples with TBS.
-
5.
Block the samples with TBS containing 10 % normal horse serum for 1 h.
-
6.
Apply the primary antibody against an endogenous nuclear body marker protein (see Note 8 ) for 1 h at room temperature or overnight at 4 °C.
-
7.
Wash the samples three times with TBST for 5 min each.
-
8.
Apply the secondary antibody conjugated to Alexa Fluor 568 for 1 h at room temperature.
-
9.
Wash the samples three times with TBST for 5 min each.
-
10.
Mount the coverslips onto slides using the VECTASHIELD Mounting Medium with DAPI.
-
11.
Visualize samples at room temperature using a fluorescence microscope (FluoView FV1000D IX81; Olympus) equipped with a U-Plan Apochromat 40×/0.95 objective lens (Olympus) (see Note 15 ).
-
12.
Acquire and process images using FluoView FV10-ASW1.7 software (Olympus) (Fig. 2).
4 Notes
-
1.
The HGPD is available at the following website: http://hgpd.lifesciencedb.jp/.
-
2.
The HGPD contains images of 15,586 N-terminal Venus fusion proteins and 17,065 C-terminal Venus fusion proteins.
-
3.
The FLJ clones that express Venus-tagged proteins that localize to nuclear foci have been listed. This list is available at the following website: http://download.hgpd.jp. The localization images of each of these proteins are accessible from the HGPD. The FLJ clones that are used for colocalization screening can be narrowed down by examining the size, shape, and number of nuclei foci in these images (Table 1, Fig. 1).
-
4.
The original Gateway Entry clones are available from the Biological Resource Center, National Institute of Technology and Evaluation (http://www.nbrc.nite.go.jp/e/hgentry-e.html). The Gateway destination vectors used to construct the expression clones of Venus-tagged proteins are available from the authors upon request.
-
5.
Although the protocol described here uses HeLa cells, it is applicable to other cultured cell lines (e.g., A549, HEK293, and NIH3T3). These cell lines are available from several cell stock centers, such as the American Type Culture Collection (http://www.atcc.org/CulturesandProducts/CellBiology/CellLinesandHybridomas/tabid/169/Default.aspx).
-
6.
Culture medium should be chosen according to the cell line that is used.
-
7.
This solution should be freshly prepared.
-
8.
The primary antibody recognizes an endogenous nuclear body marker protein. To screen for paraspeckle proteins [22], a mouse monoclonal antibody against SFPQ (Sigma) was used, which was diluted 1/100 in TBST (Fig. 2).
-
9.
Secondary antibodies should be chosen that are conjugated to fluorescent dyes that can be visualized together with Venus fluorescence.
-
10.
In total, 436 of the human proteins encoded by the FLJ vectors potentially localize to nuclear foci. It should be noted that some Venus-tagged proteins mislocalize to nuclear bodies.
-
11.
In some cases, the N-terminal Venus fusion protein, but not the C-terminal Venus fusion protein, or vice versa, localizes to a particular set of nuclear foci. We recommend these clones are adopted as candidates for further screening.
-
12.
The nuclear foci to which the listed proteins localize can be roughly subcategorized according to their size, number, and shape. The typical features of known types of nuclear bodies can be obtained from Table 1 and the literature (e.g.,1, 2).
-
13.
Transfection conditions should be optimized by varying the concentration of Lipofectamine 2000.
-
14.
The incubation time after transfection should be optimized by monitoring Venus fluorescence. In our experience, 17 h is sufficient to detect the proper localization of the Venus-tagged protein (Fig. 2). It should be noted that longer incubation (e.g., 48 h) often results in the overproduction of the Venus-tagged protein in cells, which means its precise localization cannot be judged.
-
15.
If the appropriate FLJ clones are available, the localization of a given protein to nuclear bodies should be confirmed by screening both N-terminally and C-terminally tagged versions of the protein. Finally, the localization of the corresponding endogenous protein should be confirmed by labeling with an appropriate antibody [22].
References
Spector D (2006) Snapshot: cellular bodies. Cell 127:1071
Mao YS et al (2011) Biogenesis and function of nuclear bodies. Trends Genet 27:295–306
Boisvert FM et al (2007) The multifunctional nucleolus. Nat Rev Mol Cell Biol 8:574–585
Nizami ZF et al (2010) The Cajal body and histone locus body. Cold Spring Harb Perspect Biol 2:a000653
Lamond AI, Spector DL (2003) Nuclear speckles: a model for nuclear organelles. Nat Rev Mol Cell Biol 4:605–612
Yang L et al (2011) ncRNA- and Pc2 methylation-dependent gene relocation between nuclear structures mediates gene activation programs. Cell 147:773–788
Lallemand-Breitenbach V, de Thé H (2010) PML nuclear bodies. Cold Spring Harb Perspect Biol 2:a000661
Clemson CM et al (1996) XIST RNA paints the inactive X chromosome at interphase: evidence for a novel RNA involved in nuclear/chromosome structure. J Cell Biol 132:259–275
Valgardsdottir R et al (2005) Structural and functional characterization of noncoding repetitive RNAs transcribed in stressed human cells. Mol Biol Cell 16:2597–2604
Tripathi V et al (2010) The nuclear-retained noncoding RNA MALAT1 regulates alternative splicing by modulating SR splicing factor phosphorylation. Mol Cell 39:925–938
Chen LL, Carmichael GG (2009) Altered nuclear retention of mRNAs containing inverted repeats in human embryonic stem cells: functional role of a nuclear noncoding RNA. Mol Cell 35:467–478
Clemson CM et al (2009) An architectural role for a nuclear noncoding RNA: NEAT1 RNA is essential for the structure of paraspeckles. Mol Cell 33:717–726
Sasaki YT et al (2009) MENε/β noncoding RNAs are essential for structural integrity of nuclear paraspeckles. Proc Natl Acad Sci U S A A106:2525–2530
Sunwoo H et al (2009) MEN ε/β nuclear-retained non-coding RNAs are up-regulated upon muscle differentiation and are essential components of paraspeckles. Genome Res 19:347–359
Zheng R et al (2010) Polypurine-repeat-containing RNAs: a novel class of long non-coding RNA in mammalian cells. J Cell Sci 123:3734–3744
Audas TE et al (2012) Immobilization of proteins in the nucleolus by ribosomal intergenic spacer noncoding RNA. Mol Cell 45:147–157
Andersen JS et al (2005) Nucleolar proteome dynamics. Nature 33:77–83
Saitoh N et al (2004) Proteomic analysis of interchromatin granule clusters. Mol Biol Cell 15:3876–3890
Goshima N et al (2008) Human protein factory for converting the transcriptome into an in vitro-expressed proteome. Nat Methods 5:1011–1017
Maruyama Y et al (2009) Human gene and protein database (HGPD): a novel database presenting a large quantity of experiment-based results in human proteomics. Nucleic Acids Res 37:D762–D766
Maruyama Y et al (2012) HGPD: human gene and protein database, 2012 update. Nucleic Acids Res 40:D924–D929
Naganuma T et al (2012) Alternative 3′-end processing of long noncoding RNA initiates construction of nuclear paraspeckles. EMBO J J31:4020–4034
Acknowledgments
We thank Takao Naganuma and the members of the Hirose Laboratory for their support and discussion. We also thank Yukio Maruyama for providing the localization dataset. This work was supported by the NEXT program from the Japan Society for the Promotion of Science (JSPS).
Author information
Authors and Affiliations
Corresponding author
Editor information
Editors and Affiliations
Rights and permissions
Copyright information
© 2015 Springer Science+Business Media New York
About this protocol
Cite this protocol
Hirose, T., Goshima, N. (2015). Genome-Wide Co-Localization Screening of Nuclear Body Components Using a Fluorescently Tagged FLJ cDNA Clone Library. In: Nakagawa, S., Hirose, T. (eds) Nuclear Bodies and Noncoding RNAs. Methods in Molecular Biology, vol 1262. Humana Press, New York, NY. https://doi.org/10.1007/978-1-4939-2253-6_9
Download citation
DOI: https://doi.org/10.1007/978-1-4939-2253-6_9
Published:
Publisher Name: Humana Press, New York, NY
Print ISBN: 978-1-4939-2252-9
Online ISBN: 978-1-4939-2253-6
eBook Packages: Springer Protocols