Abstract
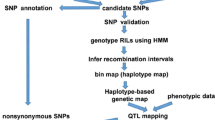
With the advancement of sequencing technologies and improvement in data analysis tools, draft genomes of many organisms are readily available. Accessibility to such draft genome sequences assists researchers, especially, plant breeders, to rapidly identify genomic regions contributing to the observed phenotypic variation leading to identification of candidate genes for a particular trait. Traditionally gene mapping is a complex, time-consuming and costly affair requiring large mapping populations and abundant molecular markers spread across the entire linkage groups. With the emergence of re-sequencing techniques, quick mapping of genes has become possible with reduced time and cost by using approaches like SHOREmap, NGM and MutMap methodologies. Among these, MutMap is widely used because it is more focused on causal SNPs. This is made possible by generating a backcross population of the mutant genotype with the parent (wild type), thereby removing the false SNPs and retaining only the SNPs linked to the mutant phenotype. Improved and specialized methods of MutMap like MutMap+, MutMap-Gap, and QTL-Seq have also emerged to expand the horizon of application of MutMap approach. The Mutmap+ methodology is specially designed for capturing those traits where the homozygous mutant leads to either lethality or sterility. MutMap-Gap methodology identifies the mutation site present in the gap regions of the reference genome, whereas QTL-Seq is an improved version of MutMap, specially designed for mapping of quantitative trait loci (QTLs). All these methods are akin to bulked segregant analysis popularly employed for mapping simply inherited traits. These methods escape the requirement of genotyping all the individuals of the mapping population and generation of high-density linkage maps for mapping of the gene for the trait of interest. This article reviews various Next Generation Sequencing-based gene mapping technologies with more emphasis on MutMap and its modifications, and discusses their advantages and proven applications for gene mapping for subsequent crop improvement.




Similar content being viewed by others
References
Abe, A., Kosugi, S., Yoshida, K., et al. (2012). Genome sequencing reveals agronomically important loci in rice using MutMap. Nature Biotechnology, 30, 174–178. https://doi.org/10.1038/nbt.2095.
Austin, R. S., Vidaurre, D., Stamatiou, G., et al. (2011). Next-generation mapping of Arabidopsis genes. Plant Journal, 67(4), 715–725. https://doi.org/10.1111/j.1365-313X.2011.04619.x.
Chen, Z., Yan, W., Wang, N., et al. (2014). Cloning of a rice male sterility gene by a modified MutMap method. Chinese, 36(1), 85–93.
Clark, A. (1976). Naturally occurring mutagens. Mutation Research, 32, 361–374.
Deng, L., Qin, P., Liu, Z., et al. (2017). Characterization and fine-mapping of a novel premature leaf senescence mutant yellow leaf and dwarf-1 in rice. Plant Physiology and Biochemistry, 111, 50–58. https://doi.org/10.1016/j.plaphy.2016.11.012.
Etherington, G. J., Monaghan, J., Zipfel, C., & MacLean, D. (2014). Mapping mutations in plant genomes with the user-friendly web application CandiSNP. Plant Methods, 10, 1–11. https://doi.org/10.1186/s13007-014-0041-7.
Fekih, R., Takagi, H., Tamiru, M., et al. (2013). MutMap+: genetic mapping and mutant identification without crossing in rice. PLoS ONE, 8, 1–10. https://doi.org/10.1371/journal.pone.0068529.
GenBank, and WGS Statistics. (2018). https://www.ncbi.nlm.nih.gov/genbank/statistics/. Accessed 18 Jul 2018.
Guo, L., Gao, Z., & Qian, Q. (2014). Application of re-sequencing to rice genomics, functional genomics and evolutionary analysis. Rice, 7, 1–10. https://doi.org/10.1186/s12284-014-0004-7.
Han, X., Xu, R., Duan, P., et al. (2017). Genetic analysis and identification of candidate genes for two spotted-leaf mutants (spl101 and spl102) in rice. Chinese, 39, 346–353. https://doi.org/10.16288/j.yczz.16-416.
Hao, N., Du, Y., Li, H., et al. (2018). CsMYB36 is involved in the formation of yellow-green peel in cucumber (Cucumis sativus L.). Theoretical and Applied Genetics, 131, 1659–1669. https://doi.org/10.1007/s00122-018-3105-7.
Henson, J., Tischler, G., & Ning, Z. (2014). Next-generation sequencing and large genome assemblies. Pharmacogenomics, 13, 901–915. https://doi.org/10.2217/pgs.12.72.
Hu, Y., Guo, L., Yang, G., et al. (2016). Genetic analysis of dense and erect panicle-2 allele DEP2-1388 and its application in hybrid rice breeding. Chinese, 38, 72–81. https://doi.org/10.16288/j.yczz.15-158.
Imamura, T., Takagi, H., Miyazato, A., et al. (2018). Isolation and characterization of the betalain biosynthesis gene involved in hypocotyl pigmentation of the allotetraploid Chenopodium quinoa. Biochemical and Biophysical Research Communications, 496, 280–286. https://doi.org/10.1016/j.bbrc.2018.01.041.
Jiao, Y., Burow, G., Gladman, N., et al. (2018). Efficient identification of causal mutations through sequencing of bulked F2 from two allelic bloomless mutants of Sorghum bicolor. Frontiers in Plant Science, 8, 1–11. https://doi.org/10.3389/fpls.2017.02267.
Lawson, N. D., & Wolfe, S. A. (2011). Forward and reverse genetic approaches for the analysis of vertebrate development in the Zebrafish. Developmental Cell, 21, 48–64. https://doi.org/10.1016/j.devcel.2011.06.007.
Liang, D., Chen, M., Qi, X., et al. (2016). QTL mapping by SLAF-seq and expression analysis of candidate genes for aphid resistance in cucumber. Frontiers in Plant Science, 7, 1–8. https://doi.org/10.3389/fpls.2016.01000.
Michelmore, R. W., Paran, I., & Kesseli, R. V. (1991). Identification of markers linked to disease-resistance genes by bulked segregant analysis: A rapid method to detect markers in specific genomic regions by using segregating populations. Proceedings of the National Academy of Sciences, 88, 9828–9832.
Nakata, M., Miyashita, T., Kimura, R., et al. (2018). MutMapPlus identified novel mutant alleles of a rice starch branching enzyme IIb gene for fine-tuning of cooked rice texture. Plant Biotechnology Journal, 16, 111–123. https://doi.org/10.1111/pbi.12753.
Oladosu, Y., Rafii, M. Y., Abdullah, N., et al. (2016). Principle and application of plant mutagenesis in crop improvement: A review. Biotechnology and Biotechnological Equipment, 30, 1–16. https://doi.org/10.1080/13102818.2015.1087333.
Pandey, M. K., Khan, A. W., Singh, V. K., et al. (2017). QTL-Seq approach identified genomic regions and diagnostic markers for rust and late leaf spot resistance in groundnut (Arachis hypogaea L.). Plant Biotechnology Journal, 15, 927–941. https://doi.org/10.1111/pbi.12686.
Pettersson, E., Lundeberg, J., & Ahmadian, A. (2009). Genomics generations of sequencing technologies. Genomics, 93, 105–111. https://doi.org/10.1016/j.ygeno.2008.10.003.
Sanger, F., Nicklen, S., & Coulson, A. R. (1977). DNA sequencing with chain-terminating inhibitors. Proceedings of the National Academy of Sciences, 74, 5463–5467. https://doi.org/10.1073/pnas.74.12.5463.
Sathya, B., Parvathy, A., & Ramesh, G. (2015). NGS meta data analysis for identification of SNP and INDEL patterns in human airway transcriptome: A preliminary indicator for lung cancer. Applied and Translational Genomics, 4, 4–9. https://doi.org/10.1016/j.atg.2014.12.003.
Schneeberger, K., Ossowski, S., Lanz, C., et al. (2009). SHOREmap: Simultaneous mapping and mutation identification by deep sequencing. Nature Methods, 6, 550–551. https://doi.org/10.1038/nmeth0809-550.
Sevanthi, A. M. V., Kandwal, P., Kale, P. B., Prakash, C., Ramkumar, M. K., Yadav, N., et al. (2018). Whole genome characterization of a few EMS-induced mutants of upland rice variety Nagina 22 reveals a staggeringly high frequency of SNPs which show high phenotypic plasticity towards the wild-type. Front Plant Sci., 9, 1179. https://doi.org/10.3389/fpls.2018.01179.
Smykal, P. (2014). Pea (Pisum sativum L.) in biology prior and after Mendel’s discovery. Czech Journal of Genetics and Plant Breeding, 50(2), 52–64. https://doi.org/10.17221/2/2014-cjgpb.
Song, J., Li, Z., Liu, Z., et al. (2017). Next-generation sequencing from bulked-segregant analysis accelerates the simultaneous identification of two qualitative genes in soybean. Frontiers in Plant Science, 8, 1–11. https://doi.org/10.3389/fpls.2017.00919.
Takagi, H., Abe, A., Yoshida, K., et al. (2013a). QTL-Seq: Rapid mapping of quantitative trait loci in rice by whole-genome resequencing of DNA from two bulked populations. Plant Journal, 74, 174–183. https://doi.org/10.1111/tpj.12105.
Takagi, H., Uemura, A., Yaegashi, H., et al. (2013b). MutMap-Gap: Whole-genome re-sequencing of mutant F2 progeny bulk combined with de novo assembly of gap regions identifies the rice blast resistance gene Pii. New Phytologist, 200, 276–283. https://doi.org/10.1111/nph.12369.
Till, B. J., Cooper, J., Tai, T. H., Colowit, P., Greene, E. A., Henikoff, S., et al. (2007). Discovery of chemically induced mutation in rice by TILLING. BMC Plant Biology, 7, 19. https://doi.org/10.1186/1471-2229-7-19.
Wadapurkar, R. M., & Vyas, R. (2018). Computational analysis of next-generation sequencing data and its applications in clinical oncology. Informatics in Medicine Unlocked, 11, 75–82. https://doi.org/10.1016/j.imu.2018.05.003.
Wang, H., Cheng, H., Wang, W., et al. (2016a). Identification of BnaYUCCA6 as a candidate gene for branch angle in Brassica napus by QTL-Seq. Scientific Reports, 6, 1–10. https://doi.org/10.1038/srep38493.
Wang, H., Li, W., Qin, Y., et al. (2017). The Cytochrome P450 Gene CsCYP85A1 is a putative candidate for super compact-1 (Scp-1) plant architecture mutation in cucumber (Cucumis sativus L.). Frontiers. Plant Science, 8, 266. https://doi.org/10.3389/fpls.2017.00266.
Wang, Y., Xiao, L., Guo, S., et al. (2016b). Fine mapping and whole-genome re-sequencing identify the seed coat color gene in Brassica rapa. PLoS ONE, 11, 1–14. https://doi.org/10.1371/journal.pone.0166464.
Xu, L., Wang, C., Cao, W., et al. (2018). CLAVATA1-type receptor-like kinase CsCLAVATA1 is a putative candidate gene for dwarf mutation in cucumber. Molecular Genetics and Genomics. https://doi.org/10.1007/s00438-018-1467-9.
Yuan, H., Fan, S., Huang, J., et al. (2017). 08SG2/OsBAK1 regulates grain size and number, and functions differently in Indica and Japonica backgrounds in rice. Rice, 10, 25. https://doi.org/10.1186/s12284-017-0165-2.
Zhang, J., Chiodini, R., Badr, A., & Zhang, G. (2011). The impact of next-generation sequencing on genomics. Journal of Genetics and Genomics, 38, 95–109. https://doi.org/10.1016/j.jgg.2011.02.003.
Zou, T., Xiao, Q., Li, W., et al. (2017). OsLAP6/OsPKS1, an orthologue of Arabidopsis PKSA/LAP6, is critical for proper pollen exine formation. Rice, 10, 53. https://doi.org/10.1186/s12284-017-0191-0.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The author declares that they have no conflict of interest.
Rights and permissions
About this article
Cite this article
Tribhuvan, K.U., Sandhya, Kumar, K. et al. MutMap: a versatile tool for identification of mutant loci and mapping of genes. Ind J Plant Physiol. 23, 612–621 (2018). https://doi.org/10.1007/s40502-018-0417-1
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s40502-018-0417-1