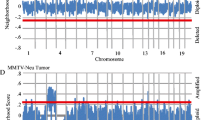
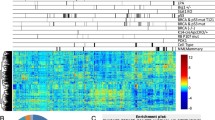
Abstract
Human breast cancer is a heterogeneous disease with numerous subtypes that have been defined through immunohistological, histological, and gene expression patterns. The diversity of breast cancer has made the study of its various underlying causes complex. To facilitate the examination of particular facets of breast cancer, mouse models have been generated, ranging from carcinogen induced models to genetically engineered mice. While mouse models have been generated to mimic the initiating event, including p53 loss, BRCA loss, or overexpression of HER2 / Neu / erbB2, other genomic events are often not well characterized. However, these secondary genetic events are often critical to the mouse tumor evolution, subtype, and outcome, just as they are in human breast cancer. As such, these other genomic events are a critical component of what models are chosen to study specific subtypes of human breast cancer. Here we review the genomic analyses that have been completed for various genetically engineered mouse models, how they compare to human breast cancer, and detail how this information can be used in choosing a mouse model for analysis.
Similar content being viewed by others
References
Greenough RB (1925) Varying degrees of malignancy in cancer of the breast.
Bloom HJG, Richardson WW. Histological grading and prognosis of breast cancer. Br J Cancer. 1957;11:359–77.
Perou CMCM, Sørile T, Eisen MBMB, et al. Molecular portraits of human breast tumours. Nature. 2000;406:747–52.
Hollern DP, Andrechek ER. A genomic analysis of mouse models of breast cancer reveals molecular features of mouse models and relationships to human breast cancer. Breast Cancer Res. 2014;16:R59. https://doi.org/10.1186/bcr3672.
Malhotra GK, Zhao X, Band H, Band V. Histological, molecular and functional subtypes of breast cancers. Cancer Biol Ther. 2010;10:955–60.
Tavassoli FA DP (2003) Tumours of the Breast World Heal Organ.
Li CI, Anderson BO, Daling JR, Moe RE. Trends in incidence rates of invasive lobular and ductal breast carcinoma. JAMA. 2003;289:1421.
Dieci MV, Orvieto E, Dominici M, Conte P, Guarneri V. Rare breast Cancer subtypes: histological, molecular, and clinical peculiarities. Oncologist. 2014;19:805–13.
Ellis IO, Galea M, Broughton N, Locker A, Blamey RW, Elston CW. Pathological prognostic factors in breast cancer. II. Histological type. Relationship with survival in a large study with long-term follow-up. Histopathology. 1992;20:479–89.
Morrogh M, Andrade VP, Giri D, Sakr RA, Paik W, Qin L-X, et al. Cadherin-catenin complex dissociation in lobular Neoplasia of the breast HHS public access. Breast Cancer Res Treat. 2012;132:641–52.
Ciriello G, Gatza ML, Beck AH, Wilkerson MD, Rhie SK, Pastore A, et al. Comprehensive molecular portraits of invasive lobular breast Cancer. Cell. 2015;163:506–19.
Wasif N, Maggard MA, Ko CY, Giuliano AE. Invasive lobular vs. ductal breast Cancer: a stage-matched comparison of outcomes. Ann Surg Oncol. 2010;17:1862–9.
van de Vijver MJ, He YD, van ‘t Veer LJ, et al. A gene-expression signature as a predictor of survival in breast Cancer. N Engl J Med. 2002;347:1999–2009.
Prat A, Parker JS, Karginova O, Fan C, Livasy C, Herschkowitz JI, et al. Phenotypic and molecular characterization of the claudin-low intrinsic subtype of breast cancer. Breast Cancer Res. 2010;12:R68. https://doi.org/10.1186/bcr2635.
Prat A, Pineda E, Adamo B, Galván P, Fernández A, Gaba L, et al. Clinical implications of the intrinsic molecular subtypes of breast cancer. Breast. 2015;24:S26–35.
Prat A, Cheang MCU, Martín M, Parker JS, Carrasco E, Caballero R, et al. Prognostic significance of progesterone receptor-positive tumor cells within immunohistochemically defined luminal a breast cancer. J Clin Oncol. 2013;31:203–9.
Parker JS, Mullins M, Cheung MCU, et al. Supervised risk predictor of breast cancer based on intrinsic subtypes. J Clin Oncol. 2009;27:1160–7.
Fan C, Oh DS, Wessels L, Weigelt B, Nuyten DSA, Nobel AB, et al. Concordance among gene-expression–based predictors for breast Cancer. N Engl J Med. 2006;355:560–9.
Fan C, Prat A, Parker JS, Liu Y, Carey LA, Troester MA, et al. Building prognostic models for breast cancer patients using clinical variables and hundreds of gene expression signatures. BMC Med Genet. 2011;4(3).
Prat A, Bianchini G, Thomas M, Belousov A, Cheang MCU, Koehler A, et al. Research-based PAM50 subtype predictor identifies higher responses and improved survival outcomes in HER2- positive breast cancer in the NOAH study. Clin Cancer Res. 2014;20:511–21.
Banerjee S, Reis-Filho JS, Ashley S, Steele D, Ashworth A, Lakhani SR, et al. Basal-like breast carcinomas: clinical outcome and response to chemotherapy. J Clin Pathol. 2006;59:729–35.
Prat A, Cruz C, Hoadley KA, Díez O, Perou CM, Balmaña J. Molecular features of the basal-like breast cancer subtype based on BRCA1 mutation status. Breast Cancer Res Treat. 2014;147:185–91.
Derose YS, Wang G, Lin YC, et al. Tumor grafts derived from women with breast cancer authentically reflect tumor pathology, growth, metastasis and disease outcomes. Nat Med. 2011;17:1514–20.
Zhang X, Lewis MT. Establishment of patient-derived Xenograft (PDX) models of human breast Cancer. Curr Protoc Mouse Biol. 2013;3:21–9.
Julien S, Merino-Trigo A, Lacroix L, Pocard M, Goere D, Mariani P, et al. Characterization of a large panel of patient-derived tumor xenografts representing the clinical heterogeneity of human colorectal cancer. Clin Cancer Res. 2012;18:5314–28.
du Manoir S, Orsetti B, Bras-Gonçalves R, Nguyen TT, Lasorsa L, Boissière F, et al. Breast tumor PDXs are genetically plastic and correspond to a subset of aggressive cancers prone to relapse. Mol Oncol. 2014;8:431–43.
Kuracha MR, Thomas P, Loggie BW, Govindarajan V. Patient-derived xenograft mouse models of pseudomyxoma peritonei recapitulate the human inflammatory tumor microenvironment. Cancer Med. 2016;5:711–9.
Villacorta-Martin C, Craig AJ, Villanueva A. Divergent evolutionary trajectories in transplanted tumor models. Nat Genet. 2017;49:1565–6.
Guenot D, Guérin E, Aguillon-Romain S, Pencreach E, Schneider A, Neuville A, et al. Primary tumour genetic alterations and intra-tumoral heterogeneity are maintained in xenografts of human colon cancers showing chromosome instability. J Pathol. 2006;208:643–52.
Yamagiwa K, Ichikawa K. Experimental study of the pathogenesis of carcinoma. J Cancer Res. 1918;27:123–81.
Pazos P, Lanari C, Meiss R, Charreau EH, Dosne Pasqualini C. Mammary carcinogenesis induced by N-methyl-N-nitrosourea (MNU) and medroxyprogesterone acetate (MPA) in BALB/c mice. Breast Cancer Res Treat. 1991;20:133–8.
Bonser M (1954) The Evolution of mammary cancer induced in female IF mice with minimal doses of locally acting methylcholanthrene.
Abba MC, Zhong Y, Lee J, Kil H, Lu Y, Takata Y, et al. DMBA induced mouse mammary tumors display high incidence of activating Pik3ca and loss of function Pten mutations. Oncotarget. 2016;7:64289–99.
Currier N, Solomon SE, Demicco EG, Chang DLF, Farago M, Ying H, et al. Oncogenic signaling pathways activated in DMBA-induced mouse mammary tumors. Toxicol Pathol. 2005;33:726–37.
Rehm S. Chemically induced mammary gland adenomyoepitheliomas and myoepithelial carcinomas of mice. Immunohistochemical and ultrastructural features. Am J Pathol. 1990;136:575–84.
Behbod F, Kittrell FS, LaMarca H, Edwards D, Kerbawy S, Heestand JC, et al. An intraductal human-in-mouse transplantation model mimics the subtypes of ductal carcinoma in situ. Breast Cancer Res. 2009;11:R66.
Valdez KE, Fan F, Smith W, Allred DC, Medina D, Behbod F. Human primary ductal carcinoma in situ (DCIS) subtype-specific pathology is preserved in a mouse intraductal (MIND) xenograft model. J Pathol. 2011;225:565–73.
D’Cruz CM, Gunther EJ, Boxer RB, et al. C-MYC induces mammary tumorigenesis by means of a preferred pathway involving spontaneous Kras2 mutations. Nat Med. 2001;7:235–9.
Andrechek ER, Cardiff RD, Chang JT, Gatza ML, Acharya CR, Potti A, et al. Genetic heterogeneity of Myc-induced mammary tumors reflecting diverse phenotypes including metastatic potential. Proc Natl Acad Sci U S A. 2009;106:16387–92.
Ivics ZN, Hackett PB, Plasterk RH, Izsvá Z. Molecular reconstruction of sleeping beauty, a Tc1-like transposon from fish, and its transposition in human cells its original location and promotes its reintegration else- where in the genome (Plasterk, 1996). Autonomous mem- bers of a transposon family can express an active trans- posase, the trans-acting factor for transposition, and thus are capable of transposing on their own. Nonauton Cell. 1997;91:501–10.
Drabek D, Zagoraiou L, DeWit T, Langeveld A, Roumpaki C, Mamalaki C, et al. Transposition of the Drosophila hydei Minos transposon in the mouse germ line. Genomics. 2003;81:108–11.
Ding S, Wu X, Li G, Han M, Zhuang Y, Xu T. Efficient transposition of the piggyBac (PB) transposon in mammalian cells and mice. Cell. 2005;122:473–83.
Collier LS, Carlson CM, Ravimohan S, Dupuy AJ, Largaespada DA. Cancer gene discovery in solid tumours using transposon-based somatic mutagenesis in the mouse. Nature. 2005;436:272–6.
Kas SM, De Ruiter JR, Schipper K, et al. Insertional mutagenesis identifies drivers of a novel oncogenic pathway in invasive lobular breast carcinoma. Nat Genet. 2017;49:1219–30.
Stewart TA, Pattengale PK, Leder P. Spontaneous mammary adenocarcinomas in transgenic mice that carry and express MTV/myc fusion genes. Cell. 1984;38:627–37.
Andres A-C, Schonenberger C-A, Groner B, Hennighausent L, Lemeur M, Gerlinger P. Ha-ras oncogene expression directed by a milk protein gene promoter: tissue specificity, hormonal regulation, and tumor induction in transgenic mice (whey acidic protein gene/whey acidic protein-ras transgene/Y chromosome integration/mammary gland tumors/salivary gland tumors). Dev Biol. 1987;84:1299–303.
Vassar R, Rosenberg M, Rosst S, Tyner A, Fuchs E (1989) Tissue-specific and differentiation-specific expression of a human K14 keratin gene in transgenic mice (stratified squamous epithelia), Tissue-specific and differentiation-specific expression of a human K14 keratin gene in transgenic mice.
Jackson EL, Willis N, Mercer K, Bronson RT, Crowley D, Montoya R, et al. Analysis of lung tumor initiation and progression using conditional expression of oncogenic K-ras. Genes Dev. 2001;15:3243–8.
Muller WJ, Sinn E, Pattengale PK, Wallace R, Leder P. Single-step induction of mammary adenocarcinoma in transgenic mice bearing the activated c-neu oncogene. Cell. 1988;54:105–15.
Gunther EJ, Belka GK, Wertheim GBW, Wang J, Hartman JL, Boxer RB, et al. A novel doxycycline-inducible system for the transgenic analysis of mammary gland biology. FASEB J. 2002;16:283–92.
Debies MT, Gestl SA, Mathers JL, Mikse OR, Leonard TL, Moody SE, et al. Tumor escape in a Wnt1-dependent mouse breast cancer model is enabled by p19Arf/p53 pathway lesions but not p16Ink4a loss. J Clin Invest. 2008;118:51–63.
Moody SE, Sarkisian CJ, Hahn KT, Gunther EJ, Pickup S, Dugan KD, et al. Conditional activation of Neu in the mammary epithelium of transgenic mice results in reversible pulmonary metastasis. Cancer Cell. 2002;2:451–61.
Podsypanina K, Politi K, Beverly LJ, Varmus HE. Oncogene cooperation in tumor maintenance and tumor recurrence in mouse mammary tumors induced by Myc and mutant Kras. Proc Natl Acad Sci. 2008;105:5242–7.
Demarest RM, Dahmane N, Capobianco AJ. Notch is oncogenic dominant in T-cell acute lymphoblastic leukemia. Blood. 2011;117:2901–9.
Wang X, Cunningham M, Zhang X, Tokarz S, Laraway B, Troxell M, et al. Phosphorylation regulates c-Myc’s oncogenic activity in the mammary gland. Cancer Res. 2011;71:925–36.
Andrechek ER. Amplification of the neu/erbB-2 oncogene in a mouse model of mammary tumorigenesis. Proc Natl Acad Sci. 2000;97:3444–9.
Andrechek ER, Hardy WR, Laing MA, Muller WJ (2004) Germ-line expression of an oncogenic erbB2 allele confers resistance to erbB2-induced mammary tumorigenesis.
Liu DP, Song H, Xu Y. A common gain of function of p53 cancer mutants in inducing genetic instability. Oncogene. 2010;29:949–56.
Yuan W, Stawiski E, Janakiraman V, et al. Conditional activation of Pik3ca H1047R in a knock-in mouse model promotes mammary tumorigenesis and emergence of mutations. Oncogene. 2013;3253:318–26.
Cressman VL, Backlund DC, Hicks EM, Gowen LC, Godfrey V, Koller BH. Mammary tumor formation in p53- and BRCA1-deficient mice. Cell Growth Differ. 1999;10:1–10.
Rao T, Ranger JJ, Smith HW, Lam SH, Chodosh L, Muller WJ (2014) Inducible and coupled expression of the polyomavirus middle T antigen and Cre recombinase in transgenic mice: an in vivo model for synthetic viability in mammary tumour progression. doi: https://doi.org/10.1186/bcr3603.
Ranger JJ, Levy DE, Shahalizadeh S, Hallett M, Muller WJ. Identification of a Stat3-dependent transcription regulatory network involved in metastatic progression. Cancer Res. 2009;69:6823–30.
Masuda T, Xu X, Dimitriadis EK, Lahusen T, Deng CX (2016) “DNA binding region” of BRCA1 affects genetic stability through modulating the intra-S-phase checkpoint. Int J Biol Sci 12:133–143.
Annunziato S, Kas SM, Nethe M, Yücel H, del Bravo J, Pritchard C, et al. Modeling invasive lobular breast carcinoma by CRISPR / Cas9-mediated somatic genome editing of the mammary gland. Genes Dev. 2016;30:1470–80.
Xue W, Chen S, Yin H, Tammela T, Papagiannakopoulos T, Joshi NS, et al. CRISPR-mediated direct mutation of cancer genes in the mouse liver HHS public access. Nature. 2014;514:380–4.
Platt RJ, Chen S, Zhou Y, Yim MJ, Swiech L, Kempton HR, et al. CRISPR-Cas9 Knockin mice for genome editing and Cancer modeling. Cell. 2014;159:440–55.
Gaj T, Gersbach CA, Barbas CF. ZFN, TALEN, and CRISPR/Cas-based methods for genome engineering. Trends Biotechnol. 2013;31:397–405.
Ahronian LG, Lewis BC. Using the RCAS-TVA system to model human Cancer in mice. Cold Spring Harb Protoc. 2014;2014:pdb.top069831.
Guy CT, Cardiff RD, Muller WJ. Induction of mammary tumors by expression of polyomavirus middle T oncogene: a transgenic mouse model for metastatic disease. Mol Cell Biol. 1992;12:954–61.
Golovkina TV, Prakash O, Ross SR. Endogenous mouse mammary tumor virus Mtv-17 is involved in Mtv-2-induced tumorigenesis in GR mice. Virology. 1996;218:14–22.
Andrechek ER. HER2/Neu tumorigenesis and metastasis is regulated by E2F activator transcription factors. Oncogene. 2015;34:217–25. https://doi.org/10.1038/onc.2013.540.
Jhan J-R, Andrechek ER. Stat3 accelerates Myc induced tumor formation while reducing growth rate in a mouse model of breast cancer. Oncotarget. 2016;7:65797–807.
Hollern DP, Honeysett J, Cardiff RD, Andrechek ER. The E2F transcription factors regulate tumor development and metastasis in a mouse model of metastatic breast Cancer. Mol Cell Biol. 2014;34:3229–43.
Cardiff RD, Anver MR, B a G, et al. The mammary pathology of genetically engineered mice: the consensus report and recommendations from the Annapolis meeting. Oncogene. 2000;19:968–88.
Cardiff RD, Wellings SR. The comparative pathology of human and mouse mammary glands. J Mammary Gland Biol Neoplasia. 1999;4:105–22.
Ponzo MG, Lesurf R, Petkiewicz S, O'Malley FP, Pinnaduwage D, Andrulis IL, et al. Met induces mammary tumors with diverse histologies and is associated with poor outcome and human basal breast cancer. Proc Natl Acad Sci U S A. 2009;106:12903–8.
Hollern DP, Swiatnicki MR, Andrechek ER. Histological subtypes of mouse mammary tumors reveal conserved relationships to human cancers. PLoS Genet. 2018;14. https://doi.org/10.1371/journal.pgen.1007135.
Lifsted T, Le Voyer T, Williams M, Muller W, Klein-Szanto A, Buetow KH, et al. Identification of inbred mouse strains harboring genetic modifiers of mammary tumor age of onset and metastatic progression. Int J Cancer. 1998;77:640–4.
Jeffers M, Fiscella M, Webb CP, Anver M, Koochekpour S, Vande Woude GF. The mutationally activated met receptor mediates motility and metastasis. Med Sci. 1998;95:14417–22.
Seth P, Porter D, Lahti-Domenici J, Geng Y, Richardson A, Polyak K, et al. Cellular and molecular targets of estrogen in normal human breast tissue. Cancer Res. 2002;62:4540–4.
Lukes L, Crawford NPS, Walker R, Hunter KW. The origins of breast Cancer prognostic gene expression profiles. Cancer Res. 2009;69:310–8.
Flowers M, Schroeder JA, Borowsky AD, Besselsen DG, Thomson CA, Pandey R, et al. Pilot study on the effects of dietary conjugated linoleic acid on tumorigenesis and gene expression in PyMT transgenic mice. Carcinogenesis. 2010;31:1642–9.
Eilon T, Barash I. Forced activation of Stat5 subjects mammary epithelial cells to DNA damage and preferential induction of the cellular response mechanism during proliferation. J Cell Physiol. 2011;226:616–26.
Lou Y, Preobrazhenska O, Auf Dem Keller U, Sutcliffe M, Barclay L, McDonald PC, et al. Epithelial-Mesenchymal transition (EMT) is not sufficient for spontaneous murine breast cancer metastasis. Dev Dyn. 2008;237:2755–68.
Kretschmer C, Sterner-Kock A, Siedentopf F, Schoenegg W, Schlag PM, Kemmner W. Identification of early molecular markers for breast cancer. Mol Cancer. 2011;10:15.
Zhang M, Tsimelzon A, Chang C-H, Fan C, Wolff A, Perou CM, et al. Intratumoral heterogeneity in a Trp53-null mouse model of human breast Cancer. Cancer Discov. 2015;5:520–33.
McBryan J, Howlin J, Kenny PA, Shioda T, Martin F. ERalpha-CITED1 co-regulated genes expressed during pubertal mammary gland development: implications for breast cancer prognosis. Oncogene. 2007;26:6406–19.
Herschkowitz JI, Simin K, Weigman VJ, Mikaelian I, Usary J, Hu Z, et al. Identification of conserved gene expression features between murine mammary carcinoma models and human breast tumors. Genome Biol. 2007;8:R76.
Slamon DJ, Clark GM, Wong SG, Levin WJ, Ullrich A, Mcguire WL. Human breast Cancer: correlation of relapse and survival with amplification of the HER-2lneu oncogene. Science (80- ). 1987;235:177–82.
Kirouac DC, Du J, Lahdenranta J, Onsum MD, Nielsen UB, Schoeberl B, et al. HER2+ Cancer cell dependence on PI3K vs. MAPK signaling axes is determined by expression of EGFR, ERBB3 and CDKN1B. PLoS Comput Biol. 2016;12:1004827.
Rennhack J, To B, Wermuth H, Andrechek ER. Mouse models of breast cancer share amplification and deletion events with human breast cancer. J Mammary Gland Biol Neoplasia. 2017;22:71–84. https://doi.org/10.1007/s10911-017-9374-y.
Santarpia L. Lippman SL. El-Naggar AK Targeting the Mitogen-Activated Protein Kinase RAS-RAF Signaling Pathway in Cancer Therapy. 2012. https://doi.org/10.1517/14728222.2011.645805.
Martini M, Chiara M, Santis D, Braccini L, Gulluni F, Hirsch E (2014) PI3K/AKT signaling pathway and cancer: an updated review. doi: https://doi.org/10.3109/07853890.2014.912836org/10.3109/07853890.2014.912836.
Bild AH, Yao G, Chang JT, Wang Q, Potti A, Chasse D, et al. Oncogenic pathway signatures in human cancers as a guide to targeted therapies. Nature. 2006;439:353–7.
Pfefferle AD, Herschkowitz JI, Usary J, Harrell J, Spike BT, Adams JR, et al. Transcriptomic classification of genetically engineered mouse models of breast cancer identifies human subtype counterparts. Genome Biol. 2013;14:R125. https://doi.org/10.1186/gb-2013-14-11-r125.
Nik-Zainal S, Davies H, Staaf J, Ramakrishna M, Glodzik D, Zou X, et al. Landscape of somatic mutations in 560 breast cancer whole-genome sequences. Nature. 2016;534:47–54.
Bose R, Kavuri SM, Searleman AC, Shen W, Shen D, Koboldt DC, et al. Activating HER2 mutations in HER2 gene amplification negative breast cancer. Cancer Discov. 2013;3:224–37.
Samuels Y, Wang Z, Bardelli A, et al. High frequency of mutations of the PIK3CA gene in human cancers. Science. 2004;80(304):554.
McFadden DG, Politi K, Bhutkar A, et al. Mutational landscape of EGFR- , MYC- , and Kras- driven genetically engineered mouse models of lung adenocarcinoma. Proc Natl Acad Sci. 2016;113:E6409–17.
Govindan R, Ding L, Griffith M, Subramanian J, Dees ND, Kanchi KL, et al. Genomic landscape of non-small cell lung cancer in smokers and never-smokers. Cell. 2012;150:1121–34.
Rennhack JP, Swiatnicki M, Zhang Y, et al Integrated sequence and gene expression analysis of mouse models of breast cancer reveals critical events with human parallels Fax Running Title: Integrated genomic characterization of mouse models of breast cancer. doi: https://doi.org/10.1101/375154
Pfefferle AD, Agrawal YN, Koboldt DC, Kanchi KL, Herschkowitz JI, Mardis ER, et al. Genomic profiling of murine mammary tumors identifies potential personalized drug targets for p53-deficient mammary cancers. Dis Model Mech. 2016;9:749–57.
Gerlinger M, Rowan AJ, Horswell S, Math M, Larkin J, Endesfelder D, et al. Intratumor heterogeneity and branched evolution revealed by multiregion sequencing. N Engl J Med. 2012;366:883–92.
Navin N, Kendall J, Troge J, Andrews P, Rodgers L, McIndoo J, et al. Tumor evolution inferred by single-cell sequencing. Nature. 2011;472:90–5.
Pal B, Chen Y, Vaillant F, Jamieson P, Gordon L, Rios AC, et al. Construction of developmental lineage relationships in the mouse mammary gland by single-cell RNA profiling. Nat Commun. 2017;8:1627.
Dai C, Arceo J, Arnold J, Sreekumar A, Dovichi NJ, Li J, Littlepage LE Metabolomics of oncogene-specific metabolic reprogramming during breast cancer. doi: https://doi.org/10.1186/s40170-018-0175-6.
Pitteri SJ, Faca VM, Kelly-Spratt KS, Kasarda AE, Wang H, Zhang Q, et al. Plasma proteome profiling of a mouse model of breast Cancer identifies a set of up-regulated proteins in common with human breast Cancer cells. J Proteome Res. 2008;7:1481–9.
Schoenherr RM, Kelly-Spratt KS, Lin C, Whiteaker JR, Liu T, Holzman T, et al. Proteome and Transcriptome profiles of a Her2/Neu-driven mouse model of breast Cancer. Proteomics Clin Appl. 2011;5:179–88.
Andrechek ER, Cardiff RD, Chang JT, Gatza ML, Acharya CR, Potti A, Nevins JR (2009) Genetic heterogeneity of Myc-induced mammary tumors reflecting diverse phenotypes including metastatic potential.
Ponzo MG, Lesurf R, Petkiewicz S, O'Malley FP, Pinnaduwage D, Andrulis IL, et al. Met induces mammary tumors with diverse histologies and is associated with poor outcome and human basal breast cancer. Proc Natl Acad Sci U S A. 2009;106:12903–8.
Fluck MM, Schaffhausen BS. Lessons in signaling and tumorigenesis from Polyomavirus middle T antigen. Microbiol Mol Biol Rev. 2009;73:542–63.
Guy CT, Webster MA, Schaller M, Parsons TJ, Cardiff RD, Muller WJ. Expression of the neu protooncogene in the mammary epithelium of transgenic mice induces metastatic disease. Proc Natl Acad Sci. 1992;89:10578–82.
Tsukamoto AS, Grosschedl R, Guzman RC, Parslow T, Varmus HE. Expression of the int-1 gene in transgenic mice is associated with mammary gland hyperplasia and adenocarcinomas in male and female mice. Cell. 1988;55:619–25.
Bocchinfuso WP, Hively WP, Couse JF, Varmus HE, Korach KS (1999) A mouse mammary tumor virus-Wnt-1 transgene induces mammary gland hyperplasia and tumorigenesis in mice lacking estrogen receptor.
Jiang Z, Deng T, Jones R, Li H, Herschkowitz JI, Liu JC, et al. Rb deletion in mouse mammary progenitors induces luminal-B or basal-like/EMT tumor subtypes depending on p53 status. J Clin Invest. 2010;120:3296–309.
Dj J, Kittrell FS, Kuperwasser C, Laucirica R, Dickinson ES, Bonilla PJ, et al. A mammary-specific model demonstrates the role of the p53 tumor suppressor gene in tumor development. Oncogene. 2000;19:1052–8.
Deng C-X, Xu X, Wagner K-U, Larson D, Weaver Z, Li C, et al. Conditional mutation of Brca1 in mammary epithelial cells results in blunted ductal morphogenesis and tumour formation. Nat Genet. 1999;22:37–43.
Chan SR, Vermi W, Luo J, Lucini L, Rickert C, Fowler AM, et al. STAT1-deficient mice spontaneously develop estrogen receptor α-positive luminal mammary carcinomas. Breast Cancer Res. 2012;14:R16.
Meraz MA, White JM, Sheehan KCF, Bach EA, Rodig SJ, Dighe AS, et al. Targeted disruption of the Stat1 gene in mice reveals unexpected physiologic specificity in the JAK-STAT signaling pathway. Cell. 1996;84:431–42.
Maroulakou IG, Anver M, Garrett L, Green JE. Prostate and mammary adenocarcinoma in transgenic mice carrying a rat C3(1) simian virus 40 large tumor antigen fusion gene. Proc Natl Acad Sci. 1994;91:11236–40.
Jones RA, Campbell CI, Gunther EJ, Chodosh LA, Petrik JJ, Khokha R, et al. Transgenic overexpression of IGF-IR disrupts mammary ductal morphogenesis and induces tumor formation. Oncogene. 2007;26:1636–44.
Moorehead RA, Barnett EF, Franks SE, Campbell CI, Siwicky MD, Livingstone J, et al. Transgenic IGF-IR overexpression induces mammary tumors with basal-like characteristics, whereas IGF-IR-independent mammary tumors express a claudin-low gene signature. Oncogene. 2011;31:3298–309.
Iavnilovitch E, Groner B, Barash I. Overexpression and forced activation of stat5 in mammary gland of transgenic mice promotes cellular proliferation, enhances differentiation, and delays postlactational apoptosis. Mol Cancer Res. 2002;1:32–47.
Bultman SJ, Herschkowitz JI, Godfrey V, Gebuhr TC, Yaniv M, Perou CM, et al. Characterization of mammary tumors from Brg1 heterozygous mice. Oncogene. 2008;27:460–8.
Tremblay PJ, Pothier F, Hoang T, Tremblay G, Brownstein S, Liszauer A, et al. Transgenic mice carrying the mouse mammary tumor virus ras fusion gene: distinct effects in various tissues. Mol Cell Biol. 1989;9:854–9.
Kuraguchi M, Ohene-Baah NY, Sonkin D, Bronson RT, Kucherlapati R. Genetic mechanisms in Apc-mediated mammary tumorigenesis. PLoS Genet. 2009;5:1000367.
Liu S, Umezu-Goto M, Murph M, Lu Y, Liu W, Zhang F, et al. Expression of Autotaxin and Lysophosphatidic acid receptors increases mammary tumorigenesis, invasion, and metastases. Cancer Cell. 2009;15:539–50.
Hu C, Diévart A, Lupien M, Calvo E, Tremblay G, Jolicoeur P. Overexpression of activated murine Notch1 and Notch3 in transgenic mice blocks mammary gland development and induces mammary tumors. Am J Pathol. 2006;168:973–90.
Usary J, Zhao W, Darr D, Roberts PJ, Liu M, Balletta L, et al. Predicting drug responsiveness in human cancers using genetically engineered mice. Clin Cancer Res. 2013;19:4889–99.
Jhan J-R, Andrechek ER. Effective personalized therapy for breast cancer based on predictions of cell signaling pathway activation from gene expression analysis. Oncogene. 2017;36:3553–61.
Funding
This work was supported with NIH R01CA160514 to E.R.A.
Author information
Authors and Affiliations
Corresponding author
Additional information
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
About this article
Cite this article
Swiatnicki, M.R., Andrechek, E.R. How to Choose a Mouse Model of Breast Cancer, a Genomic Perspective. J Mammary Gland Biol Neoplasia 24, 231–243 (2019). https://doi.org/10.1007/s10911-019-09433-3
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10911-019-09433-3