Abstract
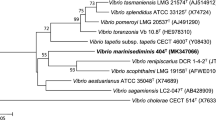
Three novel isolates (A-354T, A-328, and A-384) were retrieved from apparently healthy scleractinian Madracis decactis in the remote St Peter & St Paul Archipelago, Mid-Atlantic Ridge, Brazil. The novel isolates formed a distinct lineage based on the phylogenetic reconstruction using the 16S rRNA and pyrH gene sequences. They fell into the Mediterranei clade and their closest phylogenetic neighbour was V. mediterranei species, sharing upto 98.1 % 16S rRNA gene sequence similarity. Genomic analysis including in silico DDH, MLSA, AAI and genomic signature distinguished A-354T from V. mediterranei LMG 19703 (=AK1) with values of 33.3, 94.2, 92 %, and 11.3, respectively. Phenotypically, the novel isolates can be differentiated from V. mediterranei based on the four following features. They do not grow at 8 % NaCl; use d-gluconic acid but not l-galactonic acid lactone as carbon source; and do not have the fatty acid C18:0. Differentiation from both the other Mediterranei clade species (V. maritimus and V. variabilis) is supported by fifteen features. The novel species show lysine decarboxylase and tryptophan deaminase, but not gelatinase and arginine dihydrolase activity; produce acetoin; use α-d-lactose, N-acetyl-d-galactosamine, myo-Inositol, d-gluconic acid, and β-hydroxy-d,l-butyric acid; and present the fatty acids C14:0 iso, C15:0 anteiso, C16:0 iso, C17:0 anteiso, and C17:1x8c . Whole-cell protein profiles, based on MALDI-TOF, showed that the isolates are not clonal and also distinguished them from the closes phylogenetic neighbors. The name Vibrio madracius sp. nov. is proposed to encompass these novel isolates. The G+C content of the type strain A-354T (=LMG 28124T=CBAS 482T) is 44.5 mol%.


Similar content being viewed by others
Abbreviations
- SPSPA:
-
St Peter & St Paul Archipelago
- MLSA:
-
Multilocus sequence analysis
- ANI:
-
Average nucleotide sequence identity
- AAI:
-
Average amino acid identity
- DDH:
-
DNA–DNA hybridization
- GGDC:
-
Genome-to-genome distance calculator
- DSMZ:
-
German collection of microorganisms and cell cultures
- FAME:
-
Fatty acid methyl ester analyses
- MALDI-TOF:
-
Matrix-assisted laser desorption/ionization time-of-flight
References
Auch AF, von Jan M, Klenk H-P, Göker M (2010) Digital DNA–DNA hybridization for microbial species delineation by means of genome-to-genome sequence comparison. Stand Genomic Sci 2(1):117
Auch AF, Klenk H-P, Göker M (2010) Standard operating procedure for calculating genome-to-genome distances based on high-scoring segment pairs. Stand Genomic Sci 2(1):142–148
Aziz RK, Bartels D, Best AA, DeJongh M, Disz T, Edwards RA, Formsma K, Gerdes S, Glass EM, Kubal M, Meyer F, Olsen GJ, Olson R, Osterman AL, Overbeek RA, McNeil LK, Paarmann D, Paczian T, Parrello B, Pusch GD, Reich C, Stevens R, Vassieva O, Vonstein V, Wilke A, Zagnitko O (2008) The RAST Server: Rapid Annotations using Subsystems Technology. BMC Genom 9(1):75
Balcazar JL, Pintado J, Planas M (2010) Vibrio hippocampi sp. nov., a new species isolated from wild seahorses (Hippocampus guttulatus). FEMS Microbiol Lett 307(1):30–34
Castro B, Pires D (2006) Reproductive biology of Madracis decactis (Lyman, 1859) (Cnidaria, Scleractinia) from southern Bahia reefs, Brazil. Arqs MN 64:19–27
Chimetto LA, Cleenwerck I, Moreira APB, Brocchi M, Willems A, De Vos P, Thompson FL (2011) Vibrio variabilis sp. nov. and Vibrio maritimus sp. nov., isolated from Palythoa caribaeorum. Int J Syst Evol Microbiol 61(12):3009–3015
Felsenstein J (1985) Confidence limits on phylogenies: an approach using the bootstrap. Evolution 39(4):783–791
Karlin S, Mrázek J, Campbell AM (1998) Codon usages in different gene classes of the Escherichia coli genome. Mol Microbiol 29(6):1341–1355
Karlin S, Mrazek J, Campbell AM (1997) Compositional biases of bacterial genomes and evolutionary implications. J Bacteriol 179(12):3899–3913
Konstantinidis KT, Tiedje JM (2005) Towards a genome-based taxonomy for prokaryotes. J Bacteriol 187(18):6258–6264
Kushmaro A, Banin E, Loya Y, Stackebrandt E, Rosenberg E (2001) Vibrio shiloi sp. nov., the causative agent of bleaching of the coral Oculina patagonica. ISME J 51(4):1383–1388
Larkin M, Blackshields G, Brown N, Chenna R, McGettigan PA, McWilliam H, Valentin F, Wallace IM, Wilm A, Lopez R (2007) Clustal W and Clustal X version 2.0. Bioinformatics 23(21):2947–2948
Migotto A. (1997) Anthozoan bleaching on the southeastern coast of Brazil in the summer of 1994. In: 6th International Conference of Coelenterate Biology, Leeuwenhorst, 1995 1997. Leeuwenhorst, pp 329–335
Moreira APB, Tonon LAC, Do Valle Cecilia PP, Alves N Jr, Amado-Filho GM, Francini-Filho RB, Paranhos R, Thompson FL (2014) Culturable heterotrophic bacteria associated with healthy and bleached scleractinian Madracis decactis and the fireworm Hermodice carunculata from the Remote St. Peter and St. Paul Archipelago, Brazil. Curr Microbiol 68(1):38–46
Neves E, Johnsson R (2009) Taxonomic revision of the southwestern Atlantic Madracis and the description of Madracis fragilis n. sp. (Scleractinia: Pocilloporidae), a new coral species from Brazil. Sci Mar 73(4):739–746
Nunes FLD, Norris RD, Knowlton N (2011) Long distance dispersal and connectivity in amphi-atlantic corals at regional and basin scales. PLoS One 6(7):e22298
Ortigosa M, Garay E, Pujalte Ma-Js (1994) Numerical taxonomy of Vibrionaceae isolated from oysters and seawater along an annual cycle. Syst Appl Microbiol 17(2):216–225
Pereira-Filho GH, Amado-Filho GM, de Moura RL, Bastos AC, Guimarães SM, Salgado LT, Francini-Filho RB, Bahia RG, Abrantes DP, Guth AZ (2011) Extensive Rhodolith beds cover the summits of southwestern atlantic ocean seamounts. J Coast Res 28(1):261–269
Pujalte M-J, Garay E (1986) Proposal of Vibrio mediterranei sp. nov.: a new marine member of the genus Vibrio. Int J Syst Evol Microbiol 36(2):278–281
Pujalte MJ, Ortiz-Conde BA, Steven SE, Esteve C, Garay E, Colwell RR (1992) Numerical taxonomy and nucleic acid studies of Vibrio mediterranei. Syst Appl Microbiol 15(1):82–91
Quail MA, Smith M, Coupland P, Otto TD, Harris SR, Connor TR, Bertoni A, Swerdlow HP, Gu Y (2012) A tale of three next generation sequencing platforms: comparison of Ion Torrent, Pacific Biosciences and Illumina MiSeq sequencers. BMC Genom 13(1):341
Saitou N, Nei M (1987) The neighbor-joining method: a new method for reconstructing phylogenetic trees. Mol Biol Evol 4(4):406–425
Sawabe T, Ogura Y, Matsumura Y, Feng G, Amin AR, Mino S, Nakagawa S, Sawabe T, Kumar R, Fukui Y (2013) Updating the Vibrio clades defined by multilocus sequence phylogeny: proposal of eight new clades, and the description of Vibrio tritonius sp. nov. Front Microbiol 4:414
Strohalm M, Kavan D, Novák P, Volný M, Havlíček V (2010) mMass 3: a cross-platform software environment for precise analysis of mass spectrometric data. Anal Chem 82(11):4648–4651
Tamura K, Peterson D, Peterson N, Stecher G, Nei M, Kumar S (2011) MEGA5: molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Mol Biol Evol 28(10):2731–2739
Thompson CC, Chimetto L, Edwards RA, Swings J, Stackebrandt E, Thompson FL (2013) Microbial genomic taxonomy. BMC Genom 14(1):913
Thompson CC, Vicente AC, Souza RC, Vasconcelos AT, Vesth T, Alves N, Ussery DW, Iida T, Thompson FL (2009) Genomic taxonomy of vibrios. BMC Evol Biol 9(1):258
Thompson F, Gevers D, Thompson C, Dawyndt P, Naser S, Hoste B, Munn C, Swings J (2005) Phylogeny and molecular identification of vibrios on the basis of multilocus sequence analysis. Appl Environ Microbiol 71(9):5107–5115
Thompson FL, Hoste B, Thompson CC, Huys G, Swings J (2001) The Coral Bleaching Vibrio shiloi Kushmaro et al. 2001 is a Later Synonym of Vibrio mediterranei Pujalte and Garay 1986. Syst Appl Microbiol 24(4):516–519
Waterhouse AM, Procter JB, Martin DM, Clamp Ml, Barton GJ (2009) Jalview Version 2, A multiple sequence alignment editor and analysis workbench. Bioinformatics 25(9):1189–1191
Wieme A, Cleenwerck I, Van Landschoot A, Vandamme P (2012) Pediococcus lolii DSM 19927T and JCM 15055T are strains of Pediococcus acidilactici. Int J Syst Evol Microbiol 62(Pt 12):3105–3108
Acknowledgments
This work was supported by CNPq, CAPES, and FAPERJ. The BCCM/LMG Bacteria Collection is supported by the Federal Public Service for Science Policy, Belgium. We are grateful to Leilei Li for her assistance with the MALDI-TOF MS data analysis.
Author information
Authors and Affiliations
Corresponding author
Additional information
Nucleotide sequence data for Vibrio madracius sp. nov. are available in the DDBJ/EMBL/GenBank databases under the following accession number(s): KC751057, KC751062, KC751087 (16S rRNA); and KC751154, KC751309, KJ154030 (pyrH). ASHK00000000 is the accession number (Version ASHK00000000) for the genome of the type strain A-354T = LMG 28124T = CBAS 482 T (List in Online Resource 1). ASHJ00000000 is the accession number (Version ASHJ00000000) for the genome of V. mediterranei A-203.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Moreira, A.P.B., Duytschaever, G., Tonon, L.A.C. et al. Vibrio madracius sp. nov. Isolated from Madracis decactis (Scleractinia) in St Peter & St Paul Archipelago, Mid-Atlantic Ridge, Brazil. Curr Microbiol 69, 405–411 (2014). https://doi.org/10.1007/s00284-014-0600-1
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00284-014-0600-1