Abstract
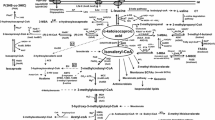
Various microorganisms, belonging to the genera Lactococcus, Lactobacillus, Streptococcus, Leuconostoc, Bifidobacterium, Propionibacterium, Brevibacterium, Corynebacterium and Arthrobacter, used in dairy fermentations such as cheese making, were analysed for their potential to convert leucine into flavour components, most notably 3-methylbutanal. A large variation between and within species was observed for various enzyme activities involved in the conversion pathway, e.g. transaminases, α-hydroxy acid dehydrogenase and α-keto acid decarboxylase. In particular, α-keto acid decarboxylase activity—leading to 3-methylbutanal—was found to be present in only two of the strains tested. It is proposed that this activity is rate-controlling in the conversion pathway leading to the flavour compound 3-methylbutanal.




Similar content being viewed by others
References
Amarita F, Fernandez Espla D, Requena T, Pelaez C (2001) Conversion of methionine to methional by Lactococcus lactis. FEMS Microbiol Lett 204:189–195
Ayad EHE, Verheul A, de Jong C, Wouters JTM, Smit G (1999) Flavour forming abilities and amino acid requirements of Lactococcus lactis strains isolated from artisanal and non-dairy origin. Int Dairy J 9:725–735
Ayad EHE, Verheul A, Wouters JTM, Smit G (2000) Application of wild starter cultures for flavour development in pilot plant cheese making. Int Dairy J 10:169–179
Bockelmann W, Hoppe Seyler T (2001) The surface flora of bacterial smear-ripened cheeses from cow’s and goat’s milk. Int Dairy J 11:307–314
Curioni PMG, Bosset JO (2002) Key odorants in various cheese types as determined by gas chromatography-olfactometry. Int Dairy J 12:959–984
Engels WJM, Visser S (1994) Isolation and comparative characterization of components that contribute to the flavour of different types of cheese. Neth Milk Dairy J 48:127–140
Engels WJM, Dekker R, de Jong C, Neeter R, Visser S (1997) A comparative study of volatile compounds in the water-soluble fraction of various types of ripened cheese. Int Dairy J 7:255–263
Engels WJM, Alting AC, Arntz MMTG, Gruppen H, Voragen AGJ, Smit G, Visser S (2000) Partial purification and characterization of two aminotransferases from Lactococcus lactis subsp. cremoris B78 involved in the catabolism of methionine and branched-chain amino acids. Int Dairy J 10:443–452
Exterkate FA, Alting AC (1990) Involvement of cell-envelope-located peptidases in the conversion of the alphas1-casein fragment f(1–23) in Gouda cheese. Brief Communications of the XXIII International Dairy Congress, Montreal, p 343
Hansen BV, Houlberg U, Ardo Y (2001) Transamination of branched-chain amino acids by a cheese related Lactobacillus paracasei strain. Int Dairy J 11:225–233
Hugenholtz J, Starrenburg M, Boels I, Sybesma W, Chaves AC, Mertens A, Kleerenbezem M (2000) Metabolic engineering of lactic acid bacteria for the improvement of fermented dairy products. In: Hofmeyr J-HS, Rohwer JM, Snoep JL (eds) Animating the cellular map. Proceedings of BTK 2000, the 9th International BioThermoKinetics meeting 3–8 April 2000. Stellenbosch University Press, South Africa, pp 3009–3013
Igoshi K (1986) Mechanism of proteolysis during cheese ripening. Jpn J Dairy Food Sci [Rakuno Kagaku Shokuhin no Kenkyu] 35:A307–A314
Jensen NBS, Melchiorsen CR, Jokumsen KV, Villadsen J (2001) Metabolic behavior of Lactococcus lactis MG1363 in microaerobic continuous cultivation at a low dilution rate. Appl Environ Microbiol 67:2677–2682
Libudzisz Z, Piatkiewicz A, Oberman H, Lubnauer M (1993) Heterogeneity of the physiological activity of Lactococcus and Leuconostoc sp. strains. Acta Microbiol Pol 42:181–192
Nierop Groot MN, de Bont JAM (1998) Conversion of phenylalanine to benzaldehyde initiated by an aminotransferase in Lactobacillus plantarum. Appl Environ Microbiol 64:3009–3013
Nosova T, Jousimies Somer H, Jokelainen K, Heine R, Salaspuro M (2000) Acetaldehyde production and metabolism by human indigenous and probiotic Lactobacillus and Bifidobacterium strains. Alcohol Alcohol 35:561–568
Oku H, Kaneda T (1988) Biosynthesis of branched-chain fatty acids in Bacillus subtilis. A decarboxylase is essential for branched-chain fatty acid synthetase. J Biol Chem 263:18386–18396
Sable S, Cottenceau G (1999) Current knowledge of soft cheeses flavor and related compounds. J Agric Food Chem 47:4825–4836
Sarkar S, Misra AK (1995) Recent trends in utilization of Propionibacterium: a review. J Dairying Food Home Sci 14:1–16
Smit G, Boven Av, Rippen M, Kruyswijk Z, Van Boven A (1998) Control of debittering activity of cheese starters. Cheese Sci 53:113
Smit G, Verheul A, van Kranenburg R, Ayad E, Siezen R, Engels W (2000) Cheese flavour development by enzymatic conversions of peptides and amino acids. Food Res Int 33:153–160
Smit G, van Hylckama Vlieg JET, Smit BA, Ayad EHE, Engels WJM (2002) Fermentative formation of flavour compounds by lactic acid bacteria. Aust J Dairy Technol 57:61–68
Ter Schure EG, Flikweert MT, Van Dijken JP, Pronk JT, Verrips CT (1998) Pyruvate decarboxylase catalyzes decarboxylation of branched-chain 2-oxo acids but is not essential for fusel alcohol production by Saccharomyces cerevisiae. Appl Environ Microbiol 64:1303–1307
Thierry A, Maillard MB, Yvon M (2002) Conversion of l-leucine to isovaleric acid by Propionibacterium freudenreichii TL 34 and ITGP23. Appl Environ Microbiol 68:608–615
Tucker JS, Morgan ME (1967) Decarboxylation of α-keto acids by Streptococcus lactis var. maltigenes. Appl Environ Microbiol 15:694–700
Van Kranenburg R, Kleerebezem M, van Hylckama Vlieg J, Ursing BM, Boekhorst J, Smit BA, Ayad EHE, Smit G, Siezen RJ (2002) Flavour formation from amino acids by lactic acid bacteria: predictions from genome sequence analysis. Int Dairy J 12:111–121
Visser S (1993) Proteolytic enzymes and their relation to cheese ripening and flavor: an overview. J Dairy Sci 76:329–350
Wallace JM, Fox PF (1997) Effect of adding free amino acids to cheddar cheese curd on proteolysis, flavour and texture development. Int Dairy J 7:157–167
Weimer B, Seefeldt K, Dias B (1999) Sulfur metabolism in bacteria associated with cheese. Antonie van Leeuwenhoek 76:247–261
Yvon M, Rijnen L (2001) Cheese flavour formation by amino acid catabolism. Int Dairy J 11:185–201
Yvon M, Berthelot S, Gripon JC (1998) Adding alpha-ketoglutarate to semi-hard cheese curd highly enhances the conversion of amino acids to aroma compounds. Int Dairy J 8:889–898
Zourari A, Accolas JP, Desmazeaud MJ (1992) Metabolism and biochemical characteristics of yogurt bacteria. A review. Lait 72:1–34
Acknowledgements
This work was financially supported by Stichting J. Mesdag Fonds, The Netherlands. The authors thank Charles Slangen and Rita Eenling for technical assistance.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Smit, B.A., Engels, W.J.M., Wouters, J.T.M. et al. Diversity of l-leucine catabolism in various microorganisms involved in dairy fermentations, and identification of the rate-controlling step in the formation of the potent flavour component 3-methylbutanal. Appl Microbiol Biotechnol 64, 396–402 (2004). https://doi.org/10.1007/s00253-003-1447-8
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00253-003-1447-8