Abstract
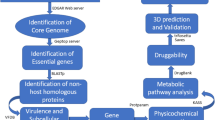
Clostridium difficile is an etiologic agent of a variety of gastrointestinal diseases in human including mild sporadic diarrhea and severe life-threatening pseudomembranous colitis. The continuous rise of C. difficile infection worldwide accompanied by rapid emergence of multidrug-resistant and hypervirulent strains has necessitated the search for novel drug targets. The present study is aimed at identifying putative therapeutic targets in this pathogen by in silico approach which encompassed four steps, viz, similarity search between pathogen and host, essentiality study using the database of essential genes, metabolic functional association study using Kyoto Encyclopedia of Genes and Genomes database, and choke point analysis. The study identified 19 promising drug targets which are non-homologous to host proteins, potentially essential for the pathogen, choke point enzymes, and participate in four pathogen-specific pathways, namely peptidoglycan biosynthesis, phosphotransferase system, two component system, and d-alanine metabolism pathways. The peptidoglycan biosynthesis pathway is the highest donor to the list of candidate target proteins. Furthermore, a three-dimensional model of one of the identified potential targets, MurG from peptidoglycan biosynthesis pathway, was constructed by homology modeling. Subsequently, by means of a virtual screening approach, the study identified eight potential inhibitors from small molecules databases, which have better docking scores, varying from −7.9 to −10.3 kcal/mol, and stronger binding affinity with target compared to known inhibitors and natural substrate of MurG. The docking analysis revealed that the active site residue Gln298 plays a critical role in ligand–target interactions which was validated through in silico mutational study. Other active site residues like Arg168, Ser198, Arg202, Ser269, and His297 were also found to play a role in binding interactions. The identified compounds may facilitate the development of new drugs to combat C. difficile-associated diseases.








Similar content being viewed by others
References
Allsop AE (1998) Bacterial genome sequencing and drug discovery. Curr Opin Biotechnol 9(6):637–642
Altschul SF, Madden TL, Schaffer AA, Zhang J, Zhang Z, Miller W, Lipman DJ (1997) Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res 25(17):3389–3402
Anishetty S, Pulimi M, Pennathur G (2005) Potential drug targets in Mycobacterium tuberculosis through metabolic pathway analysis. Comput Biol Chem 29(5):368–378
Barreteau H, Kovac A, Boniface A, Sova M, Gobec S, Blanot D (2008) Cytoplasmic steps of peptidoglycan biosynthesis. FEMS Microbiol Rev 32(2):168–207
Bartlett JG (2008) Historical perspectives on studies of Clostridium difficile and C. difficile infection. Clin Infect Dis 46(Suppl 1):S4–S11
Chhabra G, Sharma P, Anant A, Deshmukh S, Kaushik H, Gopal K, Srivastava N, Sharma N, Garg LC (2010) Identification and modeling of a drug target for Clostridium perfringens SM101. Bioinformation 4(7):278–289
Colovos C, Yeates TO (1993) Verification of protein structures: patterns of nonbonded atomic interactions. Protein Sci 2(9):1511–1519
Dutta A, Singh SK, Ghosh P, Mukherjee R, Mitter S, Bandyopadhyay D (2006) In silico identification of potential therapeutic targets in the human pathogen Helicobacter pylori. In Silico Biol 6(1–2):43–47
Eisenberg D, Luthy R, Bowie JU (1997) VERIFY3D: assessment of protein models with three-dimensional profiles. Methods Enzymol 277:396–404
El Zoeiby A, Sanschagrin F, Levesque RC (2003) Structure and function of the Mur enzymes: development of novel inhibitors. Mol Microbiol 47(1):1–12
Eswar N, John B, Mirkovic N, Fiser A, Ilyin VA, Pieper U, Stuart AC, Marti-Renom MA, Madhusudhan MS, Yerkovich B, Sali A (2003) Tools for comparative protein structure modeling and analysis. Nucleic Acids Res 31(13):3375–3380
Friesner RA, Banks JL, Murphy RB, Halgren TA, Klicic JJ, Mainz DT, Repasky MP, Knoll EH, Shelley M, Perry JK, Shaw DE, Francis P, Shenkin PS (2004) Glide: a new approach for rapid, accurate docking and scoring. 1. Method and assessment of docking accuracy. J Med Chem 47(7):1739–1749
Friesner RA, Murphy RB, Repasky MP, Frye LL, Greenwood JR, Halgren TA, Sanschagrin PC, Mainz DT (2006) Extra precision glide: docking and scoring incorporating a model of hydrophobic enclosure for protein–ligand complexes. J Med Chem 49(21):6177–6196
Galperin MY, Koonin EV (1999) Searching for drug targets in microbial genomes. Curr Opin Biotechnol 10(6):571–578
Gerding DN (2005) Metronidazole for Clostridium difficile-associated disease: is it okay for Mom? Clin Infect Dis 40(11):1598–1600
Gerding DN, Muto CA, Owens RC Jr (2008) Treatment of Clostridium difficile infection. Clin Infect Dis 46(Suppl 1):S32–S42
Goorhuis A, Van der Kooi T, Vaessen N, Dekker FW, Van den Berg R, Harmanus C, van den Hof S, Notermans DW, Kuijper EJ (2007) Spread and epidemiology of Clostridium difficile polymerase chain reaction ribotype 027/toxinotype III in The Netherlands. Clin Infect Dis 45(6):695–703
Ha S, Walker D, Shi Y, Walker S (2000) The 1.9 A crystal structure of Escherichia coli MurG, a membrane-associated glycosyltransferase involved in peptidoglycan biosynthesis. Protein Sci 9(6):1045–1052
Hu Y, Chen L, Ha S, Gross B, Falcone B, Walker D, Mokhtarzadeh M, Walker S (2003) Crystal structure of the MurG:UDP-GlcNAc complex reveals common structural principles of a superfamily of glycosyltransferases. Proc Natl Acad Sci USA 100(3):845–849
Hubert B, Loo VG, Bourgault AM, Poirier L, Dascal A, Fortin E, Dionne M, Lorange M (2007) A portrait of the geographic dissemination of the Clostridium difficile North American pulsed-field type 1 strain and the epidemiology of C. difficile-associated disease in Quebec. Clin Infect Dis 44(2):238–244
Johnson S, Gerding DN (1998) Clostridium difficile-associated diarrhea. Clin Infect Dis 26(5):1027–1034; quiz 1026–1035
Kanehisa M, Goto S, Kawashima S, Nakaya A (2002) The KEGG databases at GenomeNet. Nucleic Acids Res 30(1):42–46
Karp PD, Paley SM, Krummenacker M, Latendresse M, Dale JM, Lee TJ, Kaipa P, Gilham F, Spaulding A, Popescu L, Altman T, Paulsen I, Keseler IM, Caspi R (2009) Pathway Tools version 13.0: integrated software for pathway/genome informatics and systems biology. Brief Bioinform 11(1):40–79
Karplus M, Petsko GA (1990) Molecular dynamics simulations in biology. Nature 347(6294):631–639
Kuijper EJ, van Dissel JT, Wilcox MH (2007) Clostridium difficile: changing epidemiology and new treatment options. Curr Opin Infect Dis 20(4):376–383
Kyne L, Hamel MB, Polavaram R, Kelly CP (2002) Health care costs and mortality associated with nosocomial diarrhea due to Clostridium difficile. Clin Infect Dis 34(3):346–353
Laskowski RA, Rullmannn JA, MacArthur MW, Kaptein R, Thornton JM (1996) AQUA and PROCHECK-NMR: programs for checking the quality of protein structures solved by NMR. J Biomol NMR 8(4):477–486
Lipinski CA, Lombardo F, Dominy BW, Feeney PJ (2001) Experimental and computational approaches to estimate solubility and permeability in drug discovery and development settings. Adv Drug Deliv Rev 46(1–3):3–26
Loo VG, Poirier L, Miller MA, Oughton M, Libman MD, Michaud S, Bourgault AM, Nguyen T, Frenette C, Kelly M, Vibien A, Brassard P, Fenn S, Dewar K, Hudson TJ, Horn R, Rene P, Monczak Y, Dascal A (2005) A predominantly clonal multi-institutional outbreak of Clostridium difficile-associated diarrhea with high morbidity and mortality. N Engl J Med 353(23):2442–2449
McDonald LC, Killgore GE, Thompson A, Owens RC Jr, Kazakova SV, Sambol SP, Johnson S, Gerding DN (2005) An epidemic, toxin gene-variant strain of Clostridium difficile. N Engl J Med 353(23):2433–2441
Mooney H (2007) Annual incidence of MRSA falls in England, but C. difficile continues to rise. BMJ 335(7627):958
Ogata H, Goto S, Sato K, Fujibuchi W, Bono H, Kanehisa M (1999) KEGG: Kyoto encyclopedia of genes and genomes. Nucleic Acids Res 27(1):29–34
Perumal D, Lim CS, Sakharkar KR, Sakharkar MK (2007) Differential genome analyses of metabolic enzymes in Pseudomonas aeruginosa for drug target identification. In Silico Biol 7(4–5):453–465
Redelings MD, Sorvillo F, Mascola L (2007) Increase in Clostridium difficile-related mortality rates, United States, 1999–2004. Emerg Infect Dis 13(9):1417–1419
Rogers M (1980) Management of common skin tumours. Med J Aust 2(5):239–243
Ruiz N (2008) Bioinformatics identification of MurJ (MviN) as the peptidoglycan lipid II flippase in Escherichia coli. Proc Natl Acad Sci USA 105(40):15553–15557
Sali A, Blundell TL (1993) Comparative protein modelling by satisfaction of spatial restraints. J Mol Biol 234(3):779–815
Sarkar M, Maganti L, Ghoshal N, Dutta C (2011) In silico quest for putative drug targets in Helicobacter pylori HPAG1: molecular modeling of candidate enzymes from lipopolysaccharide biosynthesis pathway. J Mol Model 18:1855–1866
Singh S, Malik BK, Sharma DK (2007) Choke point analysis of metabolic pathways in E. histolytica: a computational approach for drug target identification. Bioinformation 2(2):68–72
Songer JG (2004) The emergence of Clostridium difficile as a pathogen of food animals. Anim Health Res Rev 5(2):321–326
Stabler RA, He M, Dawson L, Martin M, Valiente E, Corton C, Lawley TD, Sebaihia M, Quail MA, Rose G, Gerding DN, Gibert M, Popoff MR, Parkhill J, Dougan G, Wren BW (2009) Comparative genome and phenotypic analysis of Clostridium difficile 027 strains provides insight into the evolution of a hypervirulent bacterium. Genome Biol 10(9):R102
Trunkfield AE, Gurcha SS, Besra GS, Bugg TD (2010) Inhibition of Escherichia coli glycosyltransferase MurG and Mycobacterium tuberculosis Gal transferase by uridine-linked transition state mimics. Bioorg Med Chem 18(7):2651–2663
Warny M, Pepin J, Fang A, Killgore G, Thompson A, Brazier J, Frost E, McDonald LC (2005) Toxin production by an emerging strain of Clostridium difficile associated with outbreaks of severe disease in North America and Europe. Lancet 366(9491):1079–1084
Wiederstein M, Sippl MJ (2007) ProSA-web: interactive web service for the recognition of errors in three-dimensional structures of proteins. Nucleic Acids Res 35(Web Server issue):W407–W410
Yeh I, Hanekamp T, Tsoka S, Karp PD, Altman RB (2004) Computational analysis of Plasmodium falciparum metabolism: organizing genomic information to facilitate drug discovery. Genome Res 14(5):917–924
Zhang R, Lin Y (2009) DEG 5.0, a database of essential genes in both prokaryotes and eukaryotes. Nucleic Acids Res 37(Database issue):D455–D458
Acknowledgments
Vijayalakshmi is grateful to Pondicherry University, India, for the pre-doctoral fellowship. We thank Dr. Mohane Coumar, Pondicherry University, Pondicherry, for providing valuable suggestions. Research carried on at the Laboratory of the Centre for Excellence in Bioinformatics, Pondicherry University, India, is funded by the Department of Information Technology (DIT) and the Department of Biotechnology (DBT), Government of India, New Delhi, India.
Author information
Authors and Affiliations
Corresponding author
Electronic supplementary material
Below is the link to the electronic supplementary material.
44_2012_262_MOESM6_ESM.doc
Supplementary Fig. 1. 2D interaction maps for dock complexes: a–l 12 lead compounds, m known inhibitor 13b, n known inhibitor 14b and o natural substrate with target protein MurG (DOC 1121 kb)
Rights and permissions
About this article
Cite this article
Ezhilarasan, V., Sharma, O.P. & Pan, A. In silico identification of potential drug targets in Clostridium difficile R20291: modeling and virtual screening analysis of a candidate enzyme MurG. Med Chem Res 22, 2692–2705 (2013). https://doi.org/10.1007/s00044-012-0262-0
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00044-012-0262-0



