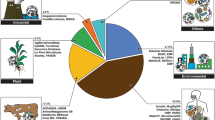
Abstract
Recent progress in the sequencing technologies and other omics approaches have had a profound impact on microbiology and helped to develop a more complete picture of the microbial composition and function of different ecosystems. One of the observations from meta-omics research is some microbes are ubiquitous in diverse ecosystems and that such shared microbiota could act as a backbone to support ecosystem function. This chapter describes current meta-omics projects studying this issue and addresses the potential of publicly available data to (i) identify the shared microbes that inhabit different environments and to (ii) study the microbial-core functions. We also discuss key challenges, gaps, and perspectives of meta-omic studies to help researchers to take the next steps forward.
Access this chapter
Tax calculation will be finalised at checkout
Purchases are for personal use only
Similar content being viewed by others
References
Abbasian F, Lockington R, Palanisami T, Ramadass K, Megharaj M, Naidu R (2016) Microbial diversity and hydrocarbon degrading gene capacity of a crude oil field soil as determined by metagenomics analysis. Biotechnol Prog 32:638–648
Abia ALK, Alisoltani A, Keshri J, Ubomba-Jaswa E (2018) Metagenomic analysis of the bacterial communities and their functional profiles in water and sediments of the Apies River, South Africa, as a function of land use. Sci Total Environ 616:326–334
Abia ALK, Alisoltani A, Ubomba-Jaswa E, Dippenaar MA (2019) Microbial life beyond the grave: 16S rRNA gene-based metagenomic analysis of bacteria diversity and their functional profiles in cemetery environments. Sci Total Environ 655:831–841
Antwis RE, Lea JM, Unwin B, Shultz S (2018) Gut microbiome composition is associated with spatial structuring and social interactions in semi-feral Welsh Mountain ponies. Microbiome 6:207
Basak P et al (2015) Spatiotemporal analysis of bacterial diversity in sediments of Sundarbans using parallel 16S rRNA gene tag sequencing. Microb Ecol 69:500–511
Bautista-de los Santos QM, Schroeder JL, Sevillano-Rivera MC, Sungthong R, Ijaz UZ, Sloan WT, Pinto AJ (2016) Emerging investigators series: microbial communities in full-scale drinking water distribution systems–a meta-analysis. Environ Sci Water Res Technol 2:631
Bell TH, Yergeau E, Juck DF, Whyte LG, Greer CW (2013a) Alteration of microbial community structure affects diesel biodegradation in an Arctic soil. FEMS Microbiol Ecol 85:51–61
Bell TH, Yergeau E, Maynard C, Juck D, Whyte LG, Greer CW (2013b) Predictable bacterial composition and hydrocarbon degradation in Arctic soils following diesel and nutrient disturbance. ISME J 7:1200–1210
Bendia AG, Signori CN, Franco DC, Duarte RT, Bohannan BJ, Pellizari VH (2018) A mosaic of geothermal and marine features shapes microbial community structure on deception Island Volcano, Antarctica. Front Microbiol 9:899
Besaury L, Ghiglione J-F, Quillet L (2014) Abundance, activity, and diversity of archaeal and bacterial communities in both uncontaminated and highly copper-contaminated marine sediments. Mar Biotechnol 16:230–242
Bolourian A, Mojtahedi Z (2018a) Immunosuppressants produced by Streptomyces: evolution, hygiene hypothesis, tumour rapalog resistance and probiotics. Environ Microbiol Rep 10:123–126
Bolourian A, Mojtahedi Z (2018b) Streptomyces, shared microbiome member of soil and gut, as ‘old friends’ against colon cancer. FEMS Microbiol Ecol 94:fiy120
Bomar L, Brugger SD, Yost BH, Davies SS, Lemon KP (2016) Corynebacterium accolens releases antipneumococcal free fatty acids from human nostril and skin surface triacylglycerols. MBio 7:e01725–e01715
Brown BL et al (2015) Metagenomic analysis of planktonic microbial consortia from a non-tidal urban-impacted segment of James River. Stand Genomic Sci 10:65
Buse HY, Lu J, Lu X, Mou X, Ashbolt NJ (2014) Microbial diversities (16S and 18S rRNA gene pyrosequencing) and environmental pathogens within drinking water biofilms grown on the common premise plumbing materials unplasticized polyvinylchloride and copper. FEMS Microbiol Ecol 88:280–295
Chao Y, Mao Y, Wang Z, Zhang T (2015) Diversity and functions of bacterial community in drinking water biofilms revealed by high-throughput sequencing. Sci Rep 5:10044
Chen SC, Musat N, Lechtenfeld OJ, Paschke H, Schmidt M, Said N, Popp D, Calabrese F, Stryhanyuk H, Jaekel U, Zhu YG (2019) Anaerobic oxidation of ethane by archaea from a marine hydrocarbon seep. Nature 568(7750):108
Chidamba L, Korsten L (2015) Pyrosequencing analysis of roof-harvested rainwater and river water used for domestic purposes in Luthengele village in the Eastern Cape Province of South Africa. Environ Monit Assess 187:1–17
Chung EJ, Park TS, Jeon CO, Chung YR (2012) Chitinophaga oryziterrae sp. nov., isolated from the rhizosphere soil of rice (Oryza sativa L.). Int J Syst Evol Microbiol 62:3030–3035
Cunha IS, Barreto CC, Costa OY, Bomfim MA, Castro AP, Kruger RH, Quirino BF (2011) Bacteria and archaea community structure in the rumen microbiome of goats (Capra hircus) from the semiarid region of Brazil. Anaerobe 17:118–124
de Macario EC, Macario AJ (2018) Methanogenic archaea in humans and other vertebrates: an update. In: (Endo) symbiotic methanogenic archaea. Springer, Berlin, pp 103–119
Deng Y, Liu P, Conrad R (2019) Effect of temperature on the microbial community responsible for methane production in alkaline NamCo wetland soil. Soil Biol Biochem 132:69–79
Dombrowski N, Donaho JA, Gutierrez T, Seitz KW, Teske AP, Baker BJ (2016) Reconstructing metabolic pathways of hydrocarbon-degrading bacteria from the Deepwater Horizon oil spill. Nat Microbiol 1:16057
Farhat M, Moletta-Denat M, Frère J, Onillon S, Trouilhé MC, Robine E (2012) Effects of disinfection on Legionella spp., eukarya, and biofilms in a hot water system. Appl Environ Microbiol 78(19):6850–6858
Flandroy L et al (2018) The impact of human activities and lifestyles on the interlinked microbiota and health of humans and of ecosystems. Sci Total Environ 627:1018–1038
Foulger R et al (2015) Representing virus-host interactions and other multi-organism processes in the gene ontology. BMC Microbiol 15:146
Francis CA, Beman JM, Kuypers MM (2007) New processes and players in the nitrogen cycle: the microbial ecology of anaerobic and archaeal ammonia oxidation. ISME J 1:19
Gosalbes MJ et al (2011) Metatranscriptomic approach to analyze the functional human gut microbiota. PLoS One 6:e17447
Grice EA et al (2009) Topographical and temporal diversity of the human skin microbiome. Science 324:1190–1192
Haggerty JM, Dinsdale EA (2017) Distinct biogeographical patterns of marine bacterial taxonomy and functional genes. Glob Ecol Biogeogr 26:177–190
Henderson G et al (2015) Rumen microbial community composition varies with diet and host, but a core microbiome is found across a wide geographical range. Sci Rep 5:14567
Hsu TK, Wu SF, Hsu BM, Kao PM, Tao CW, Shen SM, Ji WT, Huang WC, Fan CW (2015) Surveillance of parasitic Legionella in surface waters by using immunomagnetic separation and amoebae enrichment. Pathog Glob Health 109(7):328–335
Hu Y et al (2013) Metagenome-wide analysis of antibiotic resistance genes in a large cohort of human gut microbiota. Nat Commun 4:2151
Huse SM, Ye Y, Zhou Y, Fodor AA (2012) A core human microbiome as viewed through 16S rRNA sequence clusters. PLoS One 7:e34242
Huttenhower C et al (2012) Structure, function and diversity of the healthy human microbiome. Nature 486:207
Huynh HT, Pignoly M, Nkamga VD, Drancourt M, Aboudharam G (2015) The repertoire of archaea cultivated from severe periodontitis. PLoS One 10:e0121565
Jahan M, Zhanel GG, Sparling R, Holley RA (2015) Horizontal transfer of antibiotic resistance from Enterococcus faecium of fermented meat origin to clinical isolates of E. faecium and Enterococcus faecalis. Int J Food Microbiol 199:78–85
Jeon EM, Kim HJ, Jung K, Kim JH, Kim MY, Kim YP, Ka J-O (2011) Impact of Asian dust events on airborne bacterial community assessed by molecular analyses. Atmos Environ 45:4313–4321
Johnson SS, Chevrette MG, Ehlmann BL, Benison KC (2015) Insights from the metagenome of an acid salt lake: the role of biology in an extreme depositional environment. PLoS One 10:e0122869
Jordaan K, Bezuidenhout C (2016) Bacterial community composition of an urban river in the North West Province, South Africa, in relation to physico-chemical water quality. Environ Sci Pollut Res 23:5868–5880
Kamika I, Azizi S, Tekere M (2016) Microbial profiling of South African acid mine water samples using next-generation sequencing platform. Appl Microbiol Biotechnol:1–11
Kaminski J, Gibson MK, Franzosa EA, Segata N, Dantas G, Huttenhower C (2015) High-specificity targeted functional profiling in microbial communities with ShortBRED. PLoS Comput Biol 11:e1004557
Keshri J, Mankazana BB, Momba MN (2015) Profile of bacterial communities in South African mine-water samples using Illumina next-generation sequencing platform. Appl Microbiol Biotechnol 99:3233–3242
Kirs M et al (2017) Rainwater harvesting in American Samoa: current practices and indicative health risks. Environ Sci Pollut Res 24:12384–12392
Kostka JE et al (2011) Hydrocarbon-degrading bacteria and the bacterial community response in Gulf of Mexico beach sands impacted by the Deepwater Horizon oil spill. Appl Environ Microbiol 77:7962–7974
Långmark J, Storey MV, Ashbolt NJ, Stenström T-A (2005) Accumulation and fate of microorganisms and microspheres in biofilms formed in a pilot-scale water distribution system. Appl Environ Microbiol 71:706–712
Lechevallier MW, Cawthon CD, Lee RG (1988) Factors promoting survival of bacteria in chlorinated water supplies. Appl Environ Microbiol 54:649–654
Lee SA et al (2016) Comparative analysis of bacterial diversity in the rhizosphere of tomato by culture-dependent and-independent approaches. J Microbiol 54:823–831
Leewis M-C et al (2016) Differential impacts of willow and mineral fertilizer on bacterial communities and biodegradation in diesel fuel oil-contaminated soil. Front Microbiol 7
Lennard K et al (2018) Microbial composition predicts genital tract inflammation and persistent bacterial vaginosis in South African adolescent females. Infect Immun 86:e00410–e00417
Li X, Rui J, Mao Y, Yannarell A, Mackie R (2014) Dynamics of the bacterial community structure in the rhizosphere of a maize cultivar. Soil Biol Biochem 68:392–401
Lin W, Yu Z, Chen X, Liu R, Zhang H (2013) Molecular characterization of natural biofilms from household taps with different materials: PVC, stainless steel, and cast iron in drinking water distribution system. Appl Microbiol Biotechnol 97(18):8393–8401
Louca S, Parfrey LW, Doebeli M (2016) Decoupling function and taxonomy in the global ocean microbiome. Science 353:1272–1277
Lozupone CA, Stombaugh JI, Gordon JI, Jansson JK, Knight R (2012) Diversity, stability and resilience of the human gut microbiota. Nature 489:220
Ma X, Baron JL, Vikram A, Stout JE, Bibby K (2015) Fungal diversity and presence of potentially pathogenic fungi in a hospital hot water system treated with on-site monochloramine. Water Res 71:197–206
Meidute S, Demoling F, Bååth E (2008) Antagonistic and synergistic effects of fungal and bacterial growth in soil after adding different carbon and nitrogen sources. Soil Biol Biochem 40:2334–2343
Mirete S, Morgante V, González-Pastor JE (2016) Functional metagenomics of extreme environments. Curr Opin Biotechnol 38:143–149
Mitchell CM et al (2015) Colonization of the upper genital tract by vaginal bacterial species in nonpregnant women. Am J Obstet Gynecol 212:611. e611–611. e619
Moissl-Eichinger C, Probst AJ, Birarda G, Auerbach A, Koskinen K, Wolf P, Holman H-YN (2017) Human age and skin physiology shape diversity and abundance of archaea on skin. Sci Rep 7:4039
Müller O, Wilson B, Paulsen ML, Rumińska A, Armo HR, Bratbak G, Øvreås L (2018) Spatiotemporal dynamics of ammonia-oxidizing thaumarchaeota in distinct arctic water masses. Front Microbiol 9:24
Nacke H, Fischer C, Thürmer A, Meinicke P, Daniel R (2014) Land use type significantly affects microbial gene transcription in soil. Microb Ecol 67:919–930
Navarro-Noya YE, Suárez-Arriaga MC, Rojas-Valdes A, Montoya-Ciriaco NM, Gómez-Acata S, Fernández-Luqueño F, Dendooven L (2013) Pyrosequencing analysis of the bacterial community in drinking water wells. Microb Ecol 66:19–29
Nkamga VD, Henrissat B, Drancourt M (2017) Archaea: essential inhabitants of the human digestive microbiota. Hum Microbiome J 3:1–8
Oxley AP et al (2010) Halophilic archaea in the human intestinal mucosa. Environ Microbiol 12:2398–2410
Polymenakou PN (2012) Atmosphere: a source of pathogenic or beneficial microbes? Atmosphere 3:87–102
Polymenakou PN, Mandalakis M, Stephanou EG, Tselepides A (2007) Particle size distribution of airborne microorganisms and pathogens during an intense African dust event in the eastern Mediterranean. Environ Health Perspect 116:292–296
Prest E, El-Chakhtoura J, Hammes F, Saikaly P, Van Loosdrecht M, Vrouwenvelder JS (2014) Combining flow cytometry and 16S rRNA gene pyrosequencing: a promising approach for drinking water monitoring and characterization. Water Res 63:179–189
Quatrini R, Johnson DB (2018) Microbiomes in extremely acidic environments: functionalities and interactions that allow survival and growth of prokaryotes at low pH. Curr Opin Microbiol 43:139–147
Ramadass K, Megharaj M, Venkateswarlu K, Naidu R (2016) Soil bacterial strains with heavy metal resistance and high potential in degrading diesel oil and n-alkanes. Int J Environ Sci Technol 13:2863–2874
Raman R, Tharakaraman K, Sasisekharan V, Sasisekharan R (2016) Glycan–protein interactions in viral pathogenesis. Curr Opin Struct Biol 40:153–162
Ramsey MM, Freire MO, Gabrilska RA, Rumbaugh KP, Lemon KP (2016) Staphylococcus aureus shifts toward commensalism in response to Corynebacterium species. Front Microbiol 7:1230
Raymann K, Moeller AH, Goodman AL, Ochman H (2017) Unexplored archaeal diversity in the great ape gut microbiome. MSphere 2:e00026–e00017
Reese AT, Lulow K, David LA, Wright JP (2018) Plant community and soil conditions individually affect soil microbial community assembly in experimental mesocosms. Ecol Evol 8:1196–1205
Reji L, Tolar BB, Smith JM, Chavez FP, Francis CA (2019) Differential co-occurrence relationships shaping ecotype diversification within Thaumarchaeota populations in the coastal ocean water column. ISME J:1
Revetta R, Gomez‐Alvarez V, Gerke T, Santo Domingo J, Ashbolt N (2016) Changes in bacterial composition of biofilm in a metropolitan drinking water distribution system. J Appl Microbiol
Reza MS et al (2018) Metagenomic analysis using 16S ribosomal RNA genes of a bacterial community in an urban stream, the Tama River, Tokyo. Fish Sci 84:563–577
Richards CL, Broadaway SC, Eggers MJ, Doyle J, Pyle BH, Camper AK, Ford TE (2015) Detection of pathogenic and non-pathogenic bacteria in drinking water and associated biofilms on the crow reservation, Montana, USA. Microb Ecol:1–12
Rojas TCG, Lobo FP, Hongo JA, Vicentini R, Verma R, Maluta RP, da Silveira WD (2017) Genome-wide survey of genes under positive selection in avian pathogenic Escherichia coli strains. Foodborne Pathog Dis 14:245–252
Saleem F, Mustafa A, Kori JA, Hussain MS, Kamran Azim M (2018) Metagenomic characterization of bacterial communities in drinking water supply system of a mega city. Microb Ecol 76:899–910
Samad A, Trognitz F, Compant S, Antonielli L, Sessitsch A (2017) Shared and host‐specific microbiome diversity and functioning of grapevine and accompanying weed plants. Environ Microbiol 19:1407–1424
Sekar S, Zintchem AA, Keshri J, Kamika I, Momba MN (2014) Bacterial profiling in brine samples of the Emalahleni water reclamation plant, South Africa, using 454-pyrosequencing method. FEMS Microbiol Lett 359:55–63
Selden R, Widdowson P, Brooker P (2016) A reader’s guide to contemporary literary theory. Routledge, London
Serrano-Silva N, Calderon-Ezquerro MC (2018) Metagenomic survey of bacterial diversity in the atmosphere of Mexico City using different sampling methods. Environ Pollut 235:20–29
Seyler LM, McGuinness LM, Kerkhof LJ (2014) Crenarchaeal heterotrophy in salt marsh sediments. ISME J 8:1534
Shahi A, Aydin S, Ince B, Ince O (2016) Reconstruction of bacterial community structure and variation for enhanced petroleum hydrocarbons degradation through biostimulation of oil contaminated soil. Chem Eng J 306:60–66
Singh P, Jain K, Desai C, Tiwari O, Madamwar D (2019) Microbial community dynamics of extremophiles/extreme environment. In: Microbial diversity in the genomic era. Elsevier, p 323–332
Smets W, Moretti S, Denys S, Lebeer S (2016) Airborne bacteria in the atmosphere: presence, purpose, and potential. Atmos Environ 139:214–221
Steele JA et al (2011) Marine bacterial, archaeal and protistan association networks reveal ecological linkages. ISME J 5:1414
Suriya J, Shekar MC, Nathani NM, Suganya T, Bharathiraja S, Krishnan M (2017) Assessment of bacterial community composition in response to uranium levels in sediment samples of sacred Cauvery River. Appl Microbiol Biotechnol 101(2):831–841
Sutton NB et al (2013) Impact of long-term diesel contamination on soil microbial community structure. Appl Environ Microbiol 79:619–630
Tully BJ (2019) Metabolic diversity within the globally abundant marine group II Euryarchaea offers insight into ecological patterns. Nat Commun 10:271
Turnbaugh PJ et al (2009) A core gut microbiome in obese and lean twins. Nature 457:480
Violle C, Nemergut DR, Pu Z, Jiang L (2011) Phylogenetic limiting similarity and competitive exclusion. Ecol Lett 14:782–787
Wang L, Xu Q, Kong F, Yang Y, Wu D, Mishra S, Li Y (2016) Exploring the goat rumen microbiome from seven days to two years. PLoS One 11:e0154354
Wingender J, Flemming H-C (2011) Biofilms in drinking water and their role as reservoir for pathogens. Int J Hyg Environ Health 214:417–423
Wirth R et al (2018) The planktonic core microbiome and core functions in the cattle rumen by next generation sequencing. Front Microbiol 9:2285
Xi C, Zhang Y, Marrs CF, Ye W, Simon C, Foxman B, Nriagu J (2009) Prevalence of antibiotic resistance in drinking water treatment and distribution systems. Appl Environ Microbiol 75:5714–5718
Yang Y, Wang N, Guo X, Zhang Y, Ye B (2017) Comparative analysis of bacterial community structure in the rhizosphere of maize by high-throughput pyrosequencing. PLoS One 12:e0178425
Yu J, Kim D, Lee T (2010) Microbial diversity in biofilms on water distribution pipes of different materials. Water Sci Technol 61:163–171
Zaura E, Keijser BJ, Huse SM, Crielaard W (2009) Defining the healthy “core microbiome” of oral microbial communities. BMC Microbiol 9:259
Zhang G-Y, Zhang L-M, He J-Z, Liu F (2017) Comparison of archaeal populations in soil and their encapsulated Iron-manganese nodules in four locations spanning from north to South China. Geomicrobiol J 34:811–822
Zhou R, Zeng S, Hou D, Liu J, Weng S, He J, Huang Z (2019) Occurrence of human pathogenic bacteria carrying antibiotic resistance genes revealed by metagenomic approach: a case study from an aquatic environment. J Environ Sci
Acknowledgments
We thank Prof. Adam Godzik for assistance with editing and for his valuable comments that greatly improved the manuscript.
Author information
Authors and Affiliations
Corresponding author
Editor information
Editors and Affiliations
Rights and permissions
Copyright information
© 2019 Springer Nature Singapore Pte Ltd.
About this chapter
Cite this chapter
Alisoltani, A., Abia, A.L.K., Bester, L. (2019). Shared Microbiome in Different Ecosystems: A Meta-Omics Perspective. In: Tripathi, V., Kumar, P., Tripathi, P., Kishore, A., Kamle, M. (eds) Microbial Genomics in Sustainable Agroecosystems. Springer, Singapore. https://doi.org/10.1007/978-981-32-9860-6_1
Download citation
DOI: https://doi.org/10.1007/978-981-32-9860-6_1
Published:
Publisher Name: Springer, Singapore
Print ISBN: 978-981-32-9859-0
Online ISBN: 978-981-32-9860-6
eBook Packages: Biomedical and Life SciencesBiomedical and Life Sciences (R0)