Abstract
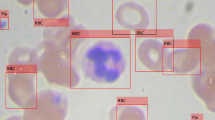
With the development of digital pathology, the automatic detection and classification of microscopy image cells using artificial intelligence technology has become a research hotspot. However, due to the problems of multi-scale cells, unbalanced foreground and background, and cell adhesion in micro images, the existing artificial intelligence object detection algorithms directly used in microscopic images have poor detection results. Therefore, to solve these problems, this paper proposes a region proposal networks based on sample weighting for cell detection and classification in microscopy images. In particular, on the basis of GA-RPN which combines the guided anchoring (GA) and the region proposal network (RPN), the sample weight learning module is added to the network, so that the network can learn the weight through the sample characteristics, which can significantly improve the detection effect. In addition, we propose a new non-maximal suppression algorithm, Density-NMS, which dynamically adjusts the suppression threshold according to the density of cell instances, allowing the model to have good detection in the case of cell adhesions. We test the proposed approach on two open cell detection challenge, Blood Cell Detection (BCCD) and Malaria. Experimental results show that the region proposal network applying sample weighting achieved superior performance on the cell detection and classification in microscopy images when compared with traditional object detection methods. In conclusion, our method can achieve better performance in object detection domain and provide an auxiliary method for pathologists to diagnose patients’ diseases.
Access this chapter
Tax calculation will be finalised at checkout
Purchases are for personal use only
Similar content being viewed by others
References
Irshad, H.: Methods for nuclei detection, segmentation, and classification in digital histopathology. IEEE Rev. Biomed. Eng. 7, 97–114 (2014)
Zhao, J., Zhang, M., Zhou, Z., Chu, J., Cao, F.: Automatic detection and classification of leukocytes using convolutional neural networks. Med. Biol. Eng. Comput. 55, 1287–1301 (2017)
Shanthi, P.B.: Deep convolution neural network for malignancy detection and classification in microscopicuterine cervix cell images. Asian Pac. J. Cancer Prevent. APJCP 20(11), 3447–3456 (2019)
Kutlu, H.: White blood cells detection and classification based on regional convolutional neural networks. Med. Hypotheses 135, 109472 (2020)
Abas, S.M.: Detection and classification of Leukocytes in Leukemia using YOLOv2 with CNN. Asian. J. Res. Comput. Sci. 8, 64–75 (2021)
Redmon, J., Divvala, S., Girshick, R., Farhadi, A.: You only look once: Unified, real-time object detection. In: 2016 IEEE Conference on Computer Vision and Pattern Recognition (CVPR), pp. 779–788. IEEE, USA (2016)
Liu, W., et al.: SSD: single shot multibox detector. In: Leibe, B., Matas, J., Sebe, N., Welling, M. (eds.) ECCV 2016. LNCS, vol. 9905, pp. 21–37. Springer, Cham (2016). https://doi.org/10.1007/978-3-319-46448-0_2
Lin, T.Y.: Focal loss for dense object detection. IEEE Trans. Pattern Anal. Mach. Intell. 42(2), 318–327 (2020)
Girshick, R.: Fast R-CNN. In: 2015 IEEE International Conference on Computer Vision (ICCV), pp. 1440–1448. IEEE, Chile (2015)
Ren, S.: Faster R-CNN: Towards real-time object detection with region proposal networks. IEEE Trans. Pattern Anal. Mach. Intell. 39(6), 1137–1149 (2020)
He, K., Gkioxari, G., Dollár, P., Girshick, R.: Mask R-CNN. In: 2017 IEEE International Conference on Computer Vision (ICCV), pp. 2980–2988. IEEE, Italy (2017)
Redmon, J., Farhadi, A.: YOLO9000: better, faster, stronger. In: 2017 IEEE Conference on Computer Vision and Pattern Recognition (CVPR), pp. 7263–7271. IEEE, USA (2017)
Yang, T., Zhang, X., Li, Z., Zhang, W., Sun, J.: Metaanchor: Learning to detect objects with customized anchors. In: Advances in Neural Information Processing Systems 31. NeurIPS, China (2018)
Zhong, Y., Wang, J., Peng, J., Zhang, L.: Anchor box optimization for object detection. In: 2020 IEEE Winter Conference on Applications of Computer Vision (WACV), pp. 1275–1283. IEEE, USA (2020)
Shrivastava, A., Gupta, A., Girshick, R.: Training region-based object detectors with online hard example mining. In: 2016 IEEE Conference on Computer Vision and Pattern Recognition (CVPR), pp. 761–769. IEEE, USA (2016)
Mikolajczyk, K.: Scale & affine invariant interest point detectors. Int. J. Comput. Vision 60, 63–86 (2004)
Viola, P., Jones, M.: Rapid object detection using a boosted cascade of simple features. In: Proceedings of the 2001 IEEE Computer Society Conference on Computer Vision and Pattern Recognition, pp. I–I. IEEE, USA (2001)
Bodla, N., Singh, B., Chellappa, R., Davis, L. S.: Soft-NMS-improving object detection with one line of code. In: 2017 IEEE International Conference on Computer Vision (ICCV), pp. 5562–5570. IEEE, Italy (2017)
Liu, S., Huang, D., Wang, Y.: Adaptive NMS: Refining pedestrian detection in a crowd. In: 2019 IEEE/CVF Conference on Computer Vision and Pattern Recognition (CVPR), pp. 6452–6461, IEEE, USA (2019)
Wang, J., Chen, K., Yang, S., Loy, C. C., Lin, D.: Region proposal by guided anchoring. In: 2019 IEEE/CVF Conference on Computer Vision and Pattern Recognition (CVPR), pp. 2960–2969. IEEE, USA (2019)
Ljosa, V.: Annotated high-throughput microscopy image sets for validation. Nat. Methods 9(7), 637–637 (2012)
Zhao, Y.Q.: Overview of deep learning object detection methods. J. Image Graph. 25(04), 0629–0654 (2020)
Redmon, J.: YOLOV3: an incremental improvement. arXiv preprint arXiv:1804.02767 (2018)
Cai, Z., Vasconcelos, N.: Cascade R-CNN: delving into high quality object detection. In: Proceedings of the IEEE conference on computer vision and pattern recognition, pp. 6154–6162. IEEE, USA (2018)
Acknowledgements
This work was supported by the National Key RD Program of China under Grant 2019YFE0190500.
Author information
Authors and Affiliations
Corresponding author
Editor information
Editors and Affiliations
Rights and permissions
Copyright information
© 2022 The Author(s), under exclusive license to Springer Nature Singapore Pte Ltd.
About this paper
Cite this paper
Chen, M., Pan, Q., Luo, Y. (2022). Density-NMS: Cell Detection and Classification in Microscopy Images. In: Wang, Y., Ma, H., Peng, Y., Liu, Y., He, R. (eds) Image and Graphics Technologies and Applications. IGTA 2022. Communications in Computer and Information Science, vol 1611. Springer, Singapore. https://doi.org/10.1007/978-981-19-5096-4_12
Download citation
DOI: https://doi.org/10.1007/978-981-19-5096-4_12
Published:
Publisher Name: Springer, Singapore
Print ISBN: 978-981-19-5095-7
Online ISBN: 978-981-19-5096-4
eBook Packages: Computer ScienceComputer Science (R0)