Abstract
Aptamers, including naturally existing riboswitches, are single-stranded nucleic acids (DNA or RNA) and peptides molecules, which show the high affinity, selectivity, specificity, and versatility. Aptamers are selected by using the systematic evolution of ligands by exponential enrichment (SELEX) method against a particular target molecule. Aptamers can target the variety of molecules like proteins, carbohydrates, bacteria, viruses, toxins, and cancerous cells and can discriminate between closely related ligands. Aptamers used various strategies to sense the target binding like electrochemical, optical, mass-sensitive analytical technique etc. Thus, aptamers magnetizes the immense interest in the field of pathogen diagnostic and detection. In this chapter, we describe how the different strategies have updated for the isolation and selection of more efficient, novel and specific aptamers. In the last, we also provide insights about the potential aptamers used for the detection several biothreat agents like bacteria and parasites, and viruses.
You have full access to this open access chapter, Download chapter PDF
Similar content being viewed by others
Keywords
1 Introduction
Aptamers are single stranded DNA or RNA oligonucleotides with unique 3D structures that allow them to bind their targets with high affinity and specificity. The possible targets of aptamers are ranging from small organic or non-organic molecules over proteins and peptides to whole cells. They are potentially regarded as alternative to antibodies to various biological and biotechnological applications (Ellington and Szostak 1990; Pfeiffer et al. 2017). Earlier aptamers were known as artificial molecules but later found in the component of naturally known riboswitches that affect transcription or translation (Lang et al. 2007; Ospina-Villa et al. 2016). Aptamers are generated in vitro by an iterative process called SELEX (Systematic Evolution of ligands by exponential enrichment).
Aptamers have been extensively studied as biomaterials because of their use in wide applications like diagnostics, therapeutics and probe for biosensors, and development of drug delivery system and novel drugs, etc. A lot of efforts have been made to search for target specific aptamers involved in several diseases like viral infections, cancer and infectious diseases involving bacterial pathogens or parasites (Bunka and Stockley 2006; Song et al. 2012). The studies involving the use of aptamers are increasing rapidly because the aptamers based technology has been known to be more effective and trustworthy. Aptamers offer several advantages over the antibodies, which are widely used in molecular diagnostics in wide range of applications (Han et al. 2010). First advantage of using oligonucleotide-based aptamer over protein-based antibodies is their stability at higher temperature, which allows them to use in many conditions. Second advantage is the cost effective production process of aptamers over laborious and extensive process of antibodies production. Also, aptamers offer advantage of easy modifications via chemical reactions to improve their stability aspects and resistance to enzyme degradation. Other advantages include the low immunogenicity and lessor toxicity of aptamers. Another important advantage offered by aptamers over antibodies is showing high affinity and binding to some ligands like ions or small molecules, which cannot be recognized by antibodies (Jayasena 1999). Based upon these advantages aptamers are regarded as alternative to antibodies in variety of applications.
2 Different Strategies for In Vitro Isolation and Selection of Aptamers
2.1 General Technique
Aptamers are generated in vitro using a conventional method referred as SELEX (Systematic evolution of ligands by exponential enrichment). Normally, SELEX is an iterative process in which the target specific sequences are selected after the repetition of selection protocols with gradually increasing the stringency (Pereira et al. 2018). Classically, the SELEX process consists of different steps where the initiation step involves the generation of ssDNA or RNA library of 1012 to 1015 sequences. Each sequence has unique feature characterized by random region of 16–75 positions flanked by two constant regions where annealing of primer takes place. In order to get the sequences, which constitute the primers of interest, ssDNA is folded and exposed to its target. The ssDNA pool is then incubated with targets and best fitting species are collected and amplified using PCR (RT-PCR in case of RNA aptamer). To ensure the higher affinity of binders with target, reiterative rounds of SELEX are executed with increasing stringent conditions (Ulrich et al. 2004). Aptamer can bind to diverse variety of targets based upon their 3D structures. The forces which plays important role in governing the 3D structures of aptamers are Watson-Crick pairing, Hodgsteen-type pairing, hydrogen bonding, electrostatic forces, and Van der Waals forces (Mercier et al. 2017).
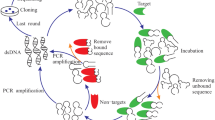
As the SELEX process involve iterations, the procedure employed to select the sequences of interests has to be repeated numerous times, and the number of cycles needed primarily depends on the rigorous conditions imposed on each cycle as well as on binding affinity between target and aptamers. Once the selection procedure is finished, an oligonucleotide population led by the target-binding sequences is obtained. The sequence of selected clones will be revealed using cloning and sequencing procedures, which further allow the prediction of the structure of selected ligands. Figure 2.1 gives the overall view of classical SELEX technology.
The initial library of ssDNA can be used instantly for the interaction with target while in case of generation of RNA library; there is an extra step for the synthesis of complementary strand by Klenow enzyme followed by in vitro transcription. Different approaches can be used to assess the interaction between target and oligonucleotides but the primary goal is to select bound species and reject the unbound ones. Once the aptamers are selected, they are PCR amplified to begin new cycle of SELEX. DNA fragments are then cloned into a plasmid and sequenced when around 90% aptamers recognize the target.
2.2 Selex Based upon Nitrocellulose Membrane Filtration
Nitrocellulose membranes primarily find their use in Western blots where they are employed to immobilize the proteins and other techniques like AFM (Atomic Force Microscopy) as these are simple and provide fast immobilization of proteins using non specific affinity of amino acids (Song et al. 2012). The SELEX method developed initially by the Gold’s group has used nitrocellulose membrane in the separation step where aptamer against T4 DNA polymerase has been developed (Tuerk and Gold 1990). During the early stages of SELEX the targets were primarily proteins therefore nitrocellulose membranes have been used for the immobilization purposes. However for small molecules they pose certain limitations as they are incapable of binding to small molecules or peptides and normally requires minimum of 12 selection rounds (Gopinath 2007).
2.3 Selex Technology Based upon Affinity Principles
Affinity chromatography techniques mainly used for the separation of recombinant proteins using specific biological affinity such as between receptor and ligand interactions or antigen-antibody interactions. Here in SELEX method, this technique is applied for the immobilization of target species on the beads and assists in the selection of library components. For the immobilization of proteins different tags like GST or His tags are used, and for small molecules targets are covalently attached to the beads using chemical reactions such as EDC coupling (Song et al. 2011). However this method has limitation, as it cannot be applied if target does not have the active functional group or affinity tag required for coupling to the beads.
Magnetic beads are also used to immobilize the targets using chemical reaction or an interaction between affinity tag and substrate on the beads. These are powerful tools for rapid isolation and selection of aptamers via immobilizing the target on magnetic beads (Joeng et al. 2009).
2.4 Selex Based upon Capillary Electrophoresis
Capillary electrophoresis (CE) offers many pleasing benefits over other separation techniques with respect to various aspects like quickness, resolution, capacity and minimum sample dilution. This technique is used to separate ionic species under the influence of electric field based upon their charge, frictional forces and hydrodynamic size (Gopinath 2007).
In this technique, aptamer can be selected among the mixture of target, library and target-library complex using mobility shift. The main advantage of this method is successful selection of the aptamer with few rounds normally two to four rounds in comparison to other methods. For example Bowser’s group constructed an aptamer for neuropeptide Y and human antibody IgE merely after four cycles of selection (Mendonsa and Bowser 2005).
2.5 Selex Based on Microfluidic Systems
To select an effective aptamer, SELEX technology using microfluidic or chip based system have been established (Hybarger et al. 2006). This technique offers advantage of efficient selection of aptamer on small scale as the method is primarily processed on a chip. For example, DNA based aptamer specific to neurotoxin type B was generated using single cycle of selection using the Continuous-flow Magnetic Activated Chip-based Separation (CMACS) device (Lou et al. 2009). The microfluidic or chip based system is grabbing attention as being an advanced method to select aptamers quickly and robotically.
2.6 Cell-SELEX Technology
Cell-SELEX is mainly designed for searching aptamers against the whole cell while the main targets of other SELEX techniques are purified proteins. The prime targets of cell- SELEX are proteins present on the extracellular surface of cell or distinctive structures specific to cell. In most cases, Cell based SELEX technology has additional steps of washing (for adhesive cells) or centrifugation (suspension cells) during the separation of aptamers as the target immobilization is not feasible in the solid phase (Fig. 2.2).
Cell SELEX uses living cells as target. In this process aptamers bind to cell membrane proteins. This procedure involves both positive selection as well as negative selection. The positive selection is incubation of target cells with library of ssDNA, followed by the collection of bound sequences. After that bound sequences are incubated with negative cell and unbound sequences are collected and served as the negative selection. The unbound sequences are used for amplification, cloning and sequencing. In this case aptamers are obtained after 10–15 alternative rounds.
Additionally, counter selections are essential in each step to avoid the non- specific aptamers, which can recognize the cell surface common to other cells. Therefore, this method is comparatively more complicated as compare to other SELEX methods because of counter selection and impossibility of immobilizing targets. However, once the aptamer selected they are powerful for cell surface diagnostics, cell specific therapy, cell targeted based drug delivery, etc. kind of different applications. For example, Kobatake’s group identified a DNA based aptamer for potential biomarker SBC3 for an adherent small lung cancer cell line (SCLC) using Cell SELEX based technology. This aptamer could be a potent SBC3 specific marker because usual biomarker (pro-GRP) which is used to diagnose SCLC is not expressed in this cell line (Kunii et al. 2011).
3 Post SELEX Optimization of Aptamers
Aptamers are widely regarded as alternative to antibodies in various applications like bio sensing, diagnostics and therapeutics. But very few aptamers have entered clinical trials, and approved by US Food and drug administration (FDA). The practical applications of aptamers are limited mainly because of instability to ribozymes, which are responsible for the degradation of aptamers by hydrolysis of phosphodiester bonds in biological environment (Gao et al. 2016). For example thrombin aptamer that showed good anticoagulant activity but has very short in vivo half-life of 108 s (Griffin et al. 1993). Consequently, it is very important to increase the thermal and nuclease stability of aptamers through modifications. Therefore, here we will highlights some of the optimization methods that have been applied in recent years to improve the stability of aptamers.
3.1 Truncation
Aptamers selected by SELEX methods usually contain a region of randomized sequences with 30–50 length nucleotides and primer sequences fixed at each end to allow the PCR amplifications (Radom et al. 2013). However, research by Ellington group suggested that constant regions in aptamers generally don’t play important role in the binding properties, and have minimum involvement in the overall structure of aptamers. Also, longer sequences add to more synthesis costs (Cowperthwaite and Ellington 2008). Additionally, Zheng findings have shown that it is useful to reduce the length of aptamers, as shorter sequences had similar or enhanced binding affinity and specificity to target in comparison to longer aptamers (Zheng et al. 2015). Also to assess the assembly of multivalent aptamers truncation become more important. In order to find which nucleotides need to be deleted, the some knowledge of conserve structure of aptamer is needed. This can be accomplished using available software algorithms like ClustalW and DNAMAN to perform multiple sequence alignment (MSA) and find the conserve high binding motif (Nadal et al. 2013). Furthermore, simulation programs such as Mfold and RNA structure can also be used to first predict the secondary structure elements, and later based upon the information of secondary structures truncation can be done in conserved stem-loop regions which likely to be involved in the binding to target. Sung group obtained the modified RNA aptamer named 2′- fluoropyrimidine (8A-W) against IL-8 (Interleukin 8) using Mfold based truncation (Sung et al. 2014).
3.2 Chemical Modifications
The chemical modifications for the optimization of aptamers for improving their stability and other aspects have been categorized into three groups: (a) modifications of sugar ring (2′ and 4′ position); (b) modifications of bases (5′ position of pyrimidine); and (c) modifications of linkage methylphosphonate and phosphorothioate replacement, 5–5′ or 3–3′ internucleotide linkage and 3′-biotin–streptavidin conjugates. RNA based aptamers are normally modified at 2′ positions of nucleosides while modifications of DNA aptamers take place at phosphodiester backbone (Wang et al. 2011).
3.3 Bivalency or Multivalency of Aptamers
The uses of monovalent aptamers are restricted in therapeutics because they offer lesser retention time of target and absence of cross-linking which may reduce their therapeutic index. Therefore, building of multivalent aptamers is an important strategy to improve the value of aptamer in clinical trials and swift detection (Mallikaratchy et al. 2010). Also, multivalent aptamers have been demonstrated to show better binding and activity over the monovalent aptamers. For example association of two thrombin binding aptamers gave ~17-fold better inhibition activity in comparison to monovalent aptamers (Kim et al. 2008). By adding the linkers, several research groups have built the bivalent or multivalent aptamers to increase the affinity as well as activity. Nonaka made a bivalent homologous aptamer through 10 mer thymine linker which have shown the binding to VEGF (Vascular Endothelial Growth Factor) with higher affinity (Kd = 30 pM) compared to monovalent aptamer (Kd = 300 pM) (Nonaka et al. 2012).
3.4 Random or Site Directed Mutagenesis
SELEX technique sometimes fails to recognize the aptamers which have high affinity to the target because of reduced library diversity due to experimental manipulations and amplification bias of PCR. Nonaka group established an efficient mutagenesis technique based on a genetic algorithm to improve the binding affinity of VEa121 (VEGF binding aptamer)(Nonaka et al. 2012). Additionally, site-directed mutagenesis can also be applied to characterize the secondary structure conformation and binding motif of aptamer (Zheng et al. 2015).
4 Role of Aptamers in the Detection of Biothreat Agents
The isolation, modification and optimization process of the aptamers sequences have established these molecules as an emerging and most promising class tool in the fields of diagnosis, therapeutics, and drug development (Wu and Kwon 2016). These molecules have proven superior in comparison to the other widely used tools and approaches (Culturing of microbes, Biochemical testing, Monoclonal antibodies, PCR amplification, and Enzyme linked assays) in the detection, inhibition and therapeutic applications. The high affinity, avidity, specificity and/or selectivity of the aptamers are the most important parameters, which are essential for the formulation of successful sensitive and specific assays for detection. Due to their great binding potential and versatility with a wide range of targets; small ions, proteins, chemicals, metabolites and whole cells, present in environment biological samples and food specimens, (Wu and Kwon 2016) various aptamers-based biosensing systems (Yang et al. 2005; Zhao et al. 2007), pharmaceutical agents and imaging probes (Nutiu and Li 2004; Yamamoto and Kumar 2000) have been isolated, identified and well-characterized (Majdinasab et al. 2018; Wu and Kwon 2016).
To date, most of the isolated aptamers against microorganisms have been selected for clinical applications and detection of environmental and food-borne pathogens using aptamers have become a promising area of research. In the following section of the current chapter, we have shed light on various reported aptamers used in the detection and/or diagnosis of several biothreat agents including bacteria, viruses, parasites, toxins and cancer cells.
4.1 Aptamers in Bacteria Detection
Due to the explosion of large human population and the emergence of several diseases causing pathogenic organisms, a rapid detection and surveillance of harmful microbes has become an important concern globally. Escherichia coli, Salmonella spp., Shigella spp., Vibrio spp., Listeria monocytogenes, Staphylococcus aureus, Bacillus cereus, Clostridium perfringens, Campylobacter jejuni, and Legionella spp., are majorly responsible for the various infectious and foodborne diseases outbreaks worldwide (Scallan et al. 2011). The infectious count of most these bacteria are as low as only ~10 colony-forming units (cfu) (Jyoti et al. 2011). Therefore, it is considerable to develop very sensitive, selective, robustic and economical strategies which are suitable for on-site detection and quantification of pathogenic bacteria and may very beneficial to prevent and control several infectious and foodborne disease (Majdinasab et al. 2018).
Major efforts made in the direction of aptamers, high-end biosensors and apta-assays development are generally based on the optical methods include colorimetry (Bayraç et al. 2017; Wu et al. 2017), surface-enhanced Raman scattering (SERS) (Duan et al. 2016a, c; Zhang et al. 2015) chemiluminescence (Hao et al. 2017a, b) fluorescence (Duan et al. 2012) and electrochemical methods (Bayraç et al. 2017; Luo et al. 2012). Although colorimetry is the most commonly employed method, it offers low sensitivity; thus, fluorescence based detection methods are quite popular due to their higher sensitivity limit as compared to other assays. In the last decades, aptamers against several bacterial species based on the different above-mentioned phenomenon have been developed and reported by various groups.
Generally, in the detection of bacteria several cell components like cell-wall members, lipopolysachharide, and outer membrane proteins various other enzymes as well as whole cells of bacteria can be used as ligands for selection and binding with aptamers (Torres-Chavolla and Alocilja 2009). Campylobacter jejuni bacterium and Sterne strain spores of Bacillus anthracis have been used as whole-cells targets for the selection of aptamers (Bruno and Kiel 1999). These developed aptamers molecules have not shown any cross-reactivity against Salmonella typhimurium, a typhoid causing bacterium, but a very little reactivity has shown in the high concentration of Helicobacter pylori and Listeria sp (Torres-Chavolla and Alocilja 2009).
Similarly, Chen et al. (2007) have also used whole cell SELEX strategy for the development of aptamer against Mycobacterium tuberculosis (H37Rv strain) (Chen et al. 2007). DNA aptamers against a large number of pathogenic bacteria which can specifically bind to the various components of the bacterial cell have been developed and reported and potentially used as prospective therapeutic agents and diagnostic tools (Bayraç et al. 2017; Brosel-Oliu et al. 2018; Bruno and Sivils 2017; Dinshaw et al. 2017; Duan et al. 2016b, c; Hao et al. 2017a; Sidhu et al. 2016; Wu et al. 2017).
A single-stranded DNA aptamer, very specific against a secreted protein, MPT64 protein of M. tuberculosis has developed form the pool of a single stranded DNA library with randomized 40-nucleotide region. The dissociation constant value (KD = 8.92 nM) of this devepoled aptmer was determined using Surface Plasmon Resonance (SPR) binding assay and the existence of conserved sequences in the aptamers and presence of most probable binding site on MPT64 protein was also determined by using several Bioinformatics analysis (Sypabekova et al. 2017).
Zelada-Guillén et al. (2009) have developed a potentiometric aptamer-based biosensor for the detection of S. enterica. They covalently immobilized the aptamer on the surface of carboxylated single-walled carbon nanotubes and based on the real -time assay. This developed aptamer is very rapid and specific, which is known as S-PS8.4, has shown no reactivity towards Escherichia coli or Lactobacillus casei (Zelada-Guillén et al. 2009).
Staphylococcus aureus, Escherichia coli, Pseudomonas aeruginose are the most prevalent bacteria involved in the sepsis and bacteremia i.e., bloodstream infections (BSI) which further lead to the vascular leakage and multiorgan failure (Cohen 2002; Dellinger et al. 2013; Rocheteau et al. 2015). But the detection of any pathogen, present in very low dose (~100 cells mL−1), is a challenging task due to the presence of various cells (RBCS, WBCS, Platelets and hemoglobin) and other constituents of blood tissue from unprocessed blood sample. Therefore, in order to lessen the time needed for BSI identification, Haijing Shen et al. (2016) have also constructed an aptamer-based capture platform for the detection of bacteria in bloodstream. Herein, they have rationally constructed a very simple aptamer based capture platform (Shen et al. 2016). They have designed a platform with a targeted aptamer that was functionalized with a mesoporous TiO2-coated magnetic nanoparticle (designated as Apt-Fe3O4@mTiO2). This developed capture platform showed a favorable bacterial-capture efficiency of about 80% even at low infectious doses (10–2000 CFU mL−1), and thus integrates the capabilities of bacterial recognition and enrichment (Cheng et al. 2015; Wang et al. 2015; Wu et al. 2014).
4.2 Aptamers in Parasites Detection
Several protozoan parasites are responsible for the millions of people deaths in the developing and underdeveloped countries. Plasmodium spp., Trypanosoma spp., Entamoeba histolytica, Leishmania spp., and Cryptosporidium parvuum are majorly causing agents of various diseases worldwide (Ospina-Villa et al. 2018). The adhesion, invasion and entry inside the host cell are most important events for the survival and pathogenesis of these intracellular parasitic organisms, and several surface components such as laminin, thrombospondin, heparin sulfate, and fibronectin molecules of the parasitic organisms are majorly contribute for the parasite-host cell interactions. Thus, the aptamers generated against these molecules, which are most-suitable detection markers, will show significant potential in the detection and identification of parasites from the bloodstream of infected individuals. Although, earlier in the field of the parasitology, only a limited number of aptamers molecules have been selected and characterized. But nowadays, a large number of DNA and RNA aptamers, which bind with the parasites or parasite-derived molecules, have resulted by the efforts of various researchers that may be used for the parasites detection and diagnostics (Ospina-Villa et al. 2018).
Firstly in 1999, Homann and Goringer, utilized cell-SELEX strategy to identify and select a RNA aptamer (designated aptamer 2–16) (Homann and Göringer 1999). It has shown high binding affinity (KD 60 nM) with a flagellar pocket protein (~42 kDa) of bloodstream stage of Trypanosoma brucei. As, Trypanosoma cruzi is also a blood-borne parasite, which causes Chagas sickness in humans. Trypomastigotes, an infective form of the parasite, only can be distinguished in the blood of infected people by utilizing a PCR-based technique. But a lower number of the parasite, present during the chronic phase of diseases, is very difficult to detect by this simple PCR method.
In this concern, Nagarkatti et al. (2012) followed a parasite concentration approach to facilitates the PCR-based detection methods. They utilized a whole cell-SELEX strategy and developed a serum stable RNA aptamers, which bound with the live T. cruzi (trypomastigotes stage). The developed and most effiecient aptamer (designated Apt68) by Nagarkatti, showed very high binding affinity (KD 8–25 nM) and high specificity with T. cruzi only. Apt68 did not interact with any other the insect stage like epimastigotes of T. cruzi nor with any other related trypanosomatid parasites, T. brucei and L. donovani. This behavior clearly suggested that the target of Apt68 apatmers was expressed only on T. cruzi (trypomastigotes stage) and it may be used exclusively and specifically for T. cruzi diagnostic applications (Nagarkatti et al. 2012).
Similarly, Barfod et al. (2009) also reported RNA aptamer that binds with a semiconserved region (known as DBL1α) of an erythrocyte membrane protein 1 from Plasmodium falciparum (PfEMP1) (Barfod et al. 2009). PfEMP1 protein is well known for its involvement in the adhesion of parasite with the blood vessels and erythrocytes cells of the host (Barfod et al. 2009). Since then, different reports on the characterized aptamers against Plasmodium proteins have been published. These aptamers are generated new ways for the development of new diagnostic and detection methods.
More importantly, lactate dehydrogenase (LDH) protein of the Plasmodium and other parasites has been used as a potential biomarker. Due to the presence of significant sequential differences between parasitic LDH and mammalian LDH protein, various aptamers against parasites have been discovered by different groups. Shum and Tanner (2008) have been indentified DNA aptamers (designated 2008s ssDNA aptamer) against P. falciparum, which bound with high affinity with the LDH enzyme and showed high affinity (KD 42 nM) and a 2:1 (LDH protein:aptamer) stoichiometry (Cheung et al. 2013). The crystal structure of LDH enzyme complex with aptamer showed the presence of a loop that specifically interacts with the aptamers. This loop is absent in the structure of the human LDH.
Moreover, Dirkzwager et al. (2015) developed and reported a rapid and novel diagnostic test for malaria. Herein, an aptamer captured the lactate dehydrogenase (PfLDH) biomarker of P. falciparum and the enzymatic activity of captured PfLDH was measured colorimetrically. This developed colorimetric assay i.e., an aptamer-tethered enzyme capture (APTEC) was robust and very sensitive (limit of detection = 4.9 ng mL_1) and could detect malaria in blood samples (Dirkzwager et al. 2015).
Ban’s group has reported electrochemical sensor based on the pL1 ssDNA aptamer, which also utilized and bind with the LDH enzyme of P. vivax and these sensors are capable to discriminate between malarial positive (i.e., presence of P. vivax and P. falciparum) and malarial negative samples (Lee et al. 2012). Surprisingly, pL1 was sequentially and structurally very different from the 2008s aptamer and targets the PvLDH with the involvement of many bridging water molecules with its predominant shape complementarity and it is totally different from that of 2008s (Choi and Ban 2016). Afterward, the group of Goswani reported a P38 ssDNA aptamer, which also recognized the PfLDH and have shown (KD 0.35 uM) (Jain et al. 2016).
P. falciparum is most virulent, fatal, and worsen species from other existing species of this parasite. Therefore, early detection and distinguishing other species is very important for the efficient treatment of malaria, particularly in patients, infected with P. falciparum. In this view, Cheug, clearly described the specificity of the previously reported ssDNA oligonucleotide aptamers (aptamer 2008s) against P. falciparum and could not show species discrimination in malaria patient blood samples (Cheung et al. 2018). Furthermore, on taking the advantage of a unique epitope (LISDAELEAIFDC) of PfLDH, Frith et al. (2018) have reported an very specific aptamer (designated LDHp 11 aptamer) against P. falciparum (Frith et al. 2018).
Moreover, a few groups of researchers have used SELEX method and reported several specific aptamers against Leishmania, a hemoflagelate of the Trypanosomatidae family, for its detection and diagnosis purpose. The group of Gonzalez has majorly focused on the selection and development of aptamers, used as an identification tool for L. infantum. They have selected and reported a pool of DNA aptamers, which were targeted the nuclear histone proteins of L. infantum (LiH2A and LiH3). Although, the histone proteins are highly conserved in nature, but these kinetoplastid histones have shown evolutionary divergenence at their N- and C-terminal, and this characteristic makes them a potential attractive aptamers binding targets. They have identified and described a population of ssDNA aptamer, specific for L. infantum. Among them most suitable aptamers (designated as AptLiH2A#1 and AptLiH2A#2) have shown higher binding affinity with LiH2A protein specifically. Another DNA aptamers recognize LiH3 of L. infantum have shown a potential application to detect leishmaniasis (Ramos et al. 2010).
Additionally, similar group has also described a new colloidal gold-based SELEX methodology and selected ssDNA aptamers for the detection of L. infantum. These obtained aptamers targeted the kinetoplastid membrane protein-11 of L. infantum (LiKMP-11). It is a cytoskeleton-associated protein, which participates in mobility of the flagellar structure. The specificity of the obtained ssDNA aptamers has confirmed by ELONA (Enzyme-Linked OligoNucleotide Assay), Western blot, and slot blot assays (Ramos et al. 2007).
4.3 Aptamers in Virus Detection
In the current scenario, viruses are responsible for the various dangerous diseases and outbreaks globally. An early detection and correct diagnosis of the viruses is the most critical factor for adequate treatment and prevention of the outbreak of viruses associated infections (Binning et al. 2012; González et al. 2016). At present, this is a bottleneck because most of the virus diagnostic tools and assays are still performed in the laboratories and conducted in batches. Recently, an over rise in the field of virus specific aptamers development, based on different phenomenon (colorimetric, refraction index changes, and fluorescence), has been observed. Now, a whole range of virus-specific aptamers have generated against a large number of viruses like vaccinia virus (Parekh et al. 2010), dengue virus (Fletcher et al. 2010), severe acute respiratory syndrome (SARS) (Shum and Tanner 2008), hepatitis C (Roh et al. 2011), human immunodeficiency virus (HIV) (Tombelli et al. 2005), apple stem pitting virus (Balogh et al. 2010), bovine viral diarrhea virus (Park et al. 2014), norovirus (Beier et al. 2014), rabies virus (Liang et al. 2014), hepatitis B (Suh et al. 2014), Ebola (Binning et al. 2013) and influenza (Gopinath et al. 2006; Lakshmipriya et al. 2013; Wandtke et al. 2015; Wongphatcharachai et al. 2012).
In viral diagnostics, the components targeted to detect the virus are whole virus (virion), nucleic acids and/or viral. The envelope is not targeted, as it has hardly any particular features to enable selective recognition (van den Kieboom et al. 2015). Cell surface antigens, integrase and protease proteins, heamglutannins, core antigen, and glycoproteins are the main targets for the binding of an aptamer. The glycoproteins such as hemagglutinin (HA) and neuraminidase (NA), which are involved in the host-virus interactions and present on the surface of the Influenza A virus have been used as a target for the selection of aptamers (Parekh et al. 2010). An aptamer (designated P30-10-16 aptamer) has shown the unique feature to distinguish the various closely related strains and subtypes of Influenza A. Thus, differentiation of highly pathogenic viral strains from the less virulent strains will facilitate the advancement in the field of diagnostics and therapeutics.
Similarly, a fluorescently labeled aptamers (designated HBs-A22) targeting a surface antigens (HBsAg) present on the infected cells (hepatocytes) from the Hepatitis B virus has been reported (Liu et al. 2010). The fluorescently label aptamers can be used and show the clear differences in the imaging of virus infected cells and non-infected cells in fluorescence microscopic studies. Moreover, for the detection of the Human Immunodeficiency Virus (HIV), an aptasensor, this target a trans-activator of transcription (Tat) protein that regulates HIV-1 infection in early stage has been developed (Chang et al. 1997; Mucha et al. 2002). Aditionally, a multicomponent reporter system, for the detection of serotypes of Dengue virus, that contains an inhibitory aptamer interacted with the restriction endonucleases along with a target complement/trigger system have also reported (Fletcher et al. 2010).
5 Conclusion
Conclusively, it is worth noting that to overcome and prevent the diseases, platform based on the aptamers is an encouraging strategy and it can be used appropriate diagnosis/detection, quantification and identification of pathogens. Thus, it is envisioned that in the near future this evolving technology may pave ways for the detection of biomarkers, and also find potential applications in diagnosis and therapy.
References
Balogh Z, Lautner G, Bardóczy V, Komorowska B, Gyurcsányi RE, Mészáros T (2010) Selection and versatile application of virus-specific aptamers. FASEB J 24(11):4187–4195
Barfod A, Persson T, Lindh J (2009) In vitro selection of RNA aptamers against a conserved region of the Plasmodium falciparum erythrocyte membrane protein 1. Parasitol Res 105(6):1557–1566
Bayraç C, Eyidoğan F, Öktem HA (2017) DNA aptamer-based colorimetric detection platform for Salmonella Enteritidis. Biosens Bioelectron 98:22–28
Beier R, Pahlke C, Quenzel P, Henseleit A, Boschke E, Cuniberti G, Labudde D (2014) Selection of a DNA aptamer against norovirus capsid protein VP1. FEMS Microbiol Lett 351(2):162–169
Binning JM, Leung DW, Amarasinghe G (2012) Aptamers in virology: recent advances and challenges. Front Microbiol 3:29
Binning JM, Wang T, Luthra P, Shabman RS, Borek DM, Liu G, Xu W, Leung DW, Basler CF, Amarasinghe GK (2013) Development of RNA aptamers targeting Ebola virus VP35. Biochemistry 52(47):8406–8419
Brosel-Oliu S, Ferreira R, Uria N, Abramova N, Gargallo R, Muñoz-Pascual F-X, Bratov A (2018) Novel impedimetric aptasensor for label-free detection of Escherichia coli O157: H7. Sensors Actuators B Chem 255:2988–2995
Bruno JG, Kiel JL (1999) In vitro selection of DNA aptamers to anthrax spores with electrochemiluminescence detection. Biosens Bioelectron 14(5):457–464
Bruno JG, Sivils JC (2017) Further characterization and independent validation of a DNA aptamer-quantum dot-based magnetic sandwich assay for Campylobacter. Folia Microbiol 62(6):485–490
Bunka DH, Stockley PG (2006) Aptamers come of age – at last. Nat Rev Microbiol 4(8):588–596
Chang HC, Samaniego F, Nair BC, Buonaguro L, Ensoli B (1997) HIV-1 Tat protein exits from cells via a leaderless secretory pathway and binds to extracellular matrix-associated heparan sulfate proteoglycans through its basic region. AIDS 11(12):1421–1431
Chen F, Zhou J, Luo F, Mohammed A-B, Zhang X-L (2007) Aptamer from whole-bacterium SELEX as new therapeutic reagent against virulent Mycobacterium tuberculosis. Biochem Biophys Res Commun 357(3):743–748
Cheng D, Yu M, Fu F, Han W, Li G, Xie J, Song Y, Swihart MT, Song E (2015) Dual recognition strategy for specific and sensitive detection of bacteria using aptamer-coated magnetic beads and antibiotic-capped gold nanoclusters. Anal Chem 88(1):820–825
Cheung Y-W, Kwok J, Law AW, Watt RM, Kotaka M, Tanner JA (2013) Structural basis for discriminatory recognition of Plasmodium lactate dehydrogenase by a DNA aptamer. Proc Natl Acad Sci 110(40):15967–15972
Cheung Y-W, Dirkzwager RM, Wong W-C, Cardoso J, Costa JDAN, Tanner JA (2018) Aptamer-mediated Plasmodium-specific diagnosis of malaria. Biochimie 145:131–136
Choi S-J, Ban C (2016) Crystal structure of a DNA aptamer bound to PvLDH elucidates novel single-stranded DNA structural elements for folding and recognition. Sci Rep 6:34998
Cohen J (2002) The immunopathogenesis of sepsis. Nature 420(6917):885–891
Cowperthwaite MC, Ellington AD (2008) Bioinformatic analysis of the contribution of primer sequences to aptamer structures. J Mol Evol 67(1):95–102
Dellinger RP, Levy MM, Rhodes A, Annane D, Gerlach H, Opal SM et al (2013) Surviving sepsis campaign: international guidelines for management of severe sepsis and septic shock, 2012. Intensive Care Med 39(2):165–228
Dinshaw IJ, Muniandy S, Teh SJ, Ibrahim F, Leo BF, Thong KL (2017) Development of an aptasensor using reduced graphene oxide chitosan complex to detect Salmonella. J Electroanal Chem 806:88–96
Dirkzwager RM, Kinghorn AB, Richards JS, Tanner JA (2015) APTEC: aptamer-tethered enzyme capture as a novel rapid diagnostic test for malaria. Chem Commun 51(22):4697–4700
Duan N, Wu S, Zhu C, Ma X, Wang Z, Yu Y, Jiang Y (2012) Dual-color upconversion fluorescence and aptamer-functionalized magnetic nanoparticles-based bioassay for the simultaneous detection of Salmonella typhimurium and Staphylococcus aureus. Anal Chim Acta 723:1–6
Duan N, Chang B, Zhang H, Wang Z, Wu S (2016a) Salmonella typhimurium detection using a surface-enhanced Raman scattering-based aptasensor. Int J Food Microbiol 218:38–43
Duan N, Wu S, Dai S, Gu H, Hao L, Ye H, Wang Z (2016b) Advances in aptasensors for the detection of food contaminants. Analyst 141(13):3942–3961
Duan N, Yan Y, Wu S, Wang Z (2016c) Vibrio parahaemolyticus detection aptasensor using surface-enhanced Raman scattering. Food Control 63:122–127
Ellington AD, Szostak JW (1990) In vitro selection of RNA molecules that bind specific ligands. Nature 346(6287):818–822
Fletcher SJ, Phillips LW, Milligan AS, Rodda SJ (2010) Toward specific detection of dengue virus serotypes using a novel modular biosensor. Biosens Bioelectron 26(4):1696–1700
Frith K-A, Fogel R, Goldring JD, Krause RG, Khati M, Hoppe H, Cromhout ME, Jiwaji M, Limson JL (2018) Towards development of aptamers that specifically bind to lactate dehydrogenase of Plasmodium falciparum through epitopic targeting. Malar J 17(1):191
Gao S, Zheng X, Jiao B, Wang L (2016) Post-SELEX optimization of aptamers. Anal Bioanal Chem 408(17):4567–4573
González VM, Martín ME, Fernández G, García-Sacristán A (2016) Use of aptamers as diagnostics tools and antiviral agents for human viruses. Pharmaceuticals 9(4):78
Gopinath SCB (2007) Methods developed for SELEX. Anal Bioanal Chem 387(1):171–182
Gopinath SC, Misono TS, Kawasaki K, Mizuno T, Imai M, Odagiri T, Kumar PK (2006) An RNA aptamer that distinguishes between closely related human influenza viruses and inhibits haemagglutinin-mediated membrane fusion. J Gen Virol 87(3):479–487
Griffin LC, Tidmarsh GF, Bock LC, Toole JJ, Leung L (1993) In vivo anticoagulant properties of a novel nucleotide-based thrombin inhibitor and demonstration of regional anticoagulation in extracorporeal circuits. Blood 81(12):3271–3276
Han K, Liang Z, Zhou N (2010) Design strategies for aptamer-based biosensors. Sensors 10(5):4541–4557
Hao L, Gu H, Duan N, Wu S, Ma X, Xia Y, Tao Z, Wang Z (2017a) An enhanced chemiluminescence resonance energy transfer aptasensor based on rolling circle amplification and WS2 nanosheet for Staphylococcus aureus detection. Anal Chim Acta 959:83–90
Hao L, Gu H, Duan N, Wu S, Ma X, Xia Y, Wang H, Wang Z (2017b) A chemiluminescent aptasensor based on rolling circle amplification and Co2+/N-(aminobutyl)-N-(ethylisoluminol) functional flowerlike gold nanoparticles for Salmonella typhimurium detection. Talanta 164:275–282
Homann M, Göringer HU (1999) Combinatorial selection of high affinity RNA ligands to live African trypanosomes. Nucleic Acids Res 27(9):2006–2014
Hybarger G, Bynum J, Williams RF, Valdes JJ, Chambers JP (2006) A microfluidic SELEX prototype. Anal Bioanal Chem 384(1):191–198
Jain P, Chakma B, Singh NK, Patra S, Goswami P (2016) Aromatic surfactant as aggregating agent for aptamer-gold nanoparticle-based detection of Plasmodium lactate dehydrogenase. Mol Biotechnol 58(7):497–508
Jayasena SD (1999) Aptamers: an emerging class of molecules that rival antibodies in diagnostics. Clin Chem 45(9):1628–1650
Joeng CB, Niazi JH, Lee SJ, Gu MB (2009) ssDNA aptamers that recognize diclofenac and 2-anilinophenylacetic acid. Bioorg Med Chem 17(15):5380–5387
Jyoti A, Vajpayee P, Singh G, Patel CB, Gupta KC, Shanker R (2011) Identification of environmental reservoirs of nontyphoidal salmonellosis: aptamer-assisted bioconcentration and subsequent detection of Salmonella typhimurium by quantitative polymerase chain reaction. Environ Sci Technol 45(20):8996–9002
Kim Y, Cao Z, Tan W (2008) Molecular assembly for high-performance bivalent nucleic acid inhibitor. Proc Natl Acad Sci 105(15):5664–5669
Kunii T, Ogura S-i, Mie M, Kobatake E (2011) Selection of DNA aptamers recognizing small cell lung cancer using living cell-SELEX. Analyst 136(7):1310–1312
Lakshmipriya T, Fujimaki M, Gopinath SC, Awazu K (2013) Generation of anti-influenza aptamers using the systematic evolution of ligands by exponential enrichment for sensing applications. Langmuir 29(48):15107–15115
Lang K, Rieder R, Micura R (2007) Ligand-induced folding of the thiM TPP riboswitch investigated by a structure-based fluorescence spectroscopic approach. Nucleic Acids Res 35(16):5370–5378
Lee S, Song K-M, Jeon W, Jo H, Shim Y-B, Ban C (2012) A highly sensitive aptasensor towards Plasmodium lactate dehydrogenase for the diagnosis of malaria. Biosens Bioelectron 35(1):291–296
Liang H-R, Hu G-Q, Xue X-H, Li L, Zheng X-X, Gao Y-W, Yang ST, Xia X-Z (2014) Selection of an aptamer against rabies virus: a new class of molecules with antiviral activity. Virus Res 184:7–13
Liu J, Yang Y, Hu B, Ma Z-y, Huang H-p, Yu Y, Yang D-l (2010) Development of HBsAg-binding aptamers that bind HepG2. 2.15 cells via HBV surface antigen. Virol Sin 25(1):27–35
Lou X, Qian J, Xiao Y, Viel L, Gerdon AE, Lagally ET, Soh HT (2009) Micromagnetic selection of aptamers in microfluidic channels. Proc Natl Acad Sci 106(9):2989–2994
Luo C, Lei Y, Yan L, Yu T, Li Q, Zhang D, Ju H (2012) A rapid and sensitive aptamer-based electrochemical biosensor for direct detection of Escherichia Coli O111. Electroanalysis 24(5):1186–1191
Majdinasab M, Hayat A, Marty JL (2018) Aptamer-based assays and aptasensors for detection of pathogenic bacteria in food samples. TrAC Trends Anal Chem 107:60–77
Mallikaratchy PR, Ruggiero A, Gardner JR, Kuryavyi V, Maguire WF, Heaney ML, McDevitt MR, Patel DJ, Scheinberg DA (2010) A multivalent DNA aptamer specific for the B-cell receptor on human lymphoma and leukemia. Nucleic Acids Res 39(6):2458–2469
Mendonsa SD, Bowser MT (2005) In vitro selection of aptamers with affinity for neuropeptide Y using capillary electrophoresis. J Am Chem Soc 127(26):9382–9383
Mercier M-C, Dontenwill M, Choulier L (2017) Selection of nucleic acid aptamers targeting tumor cell-surface protein biomarkers. Cancers 9(6):69
Mucha P, Szyk A, Rekowski P, Barciszewski J (2002) Structural requirements for conserved Arg52 residue for interaction of the human immunodeficiency virus type 1 trans-activation responsive element with trans-activator of transcription protein (49–57): capillary electrophoresis mobility shift assay. J Chromatogr A 968(1–2):211–220
Nadal P, Svobodova M, Mairal T, O’Sullivan C (2013) Probing high-affinity 11-mer DNA aptamer against Lup an 1 (β-conglutin). Anal Bioanal Chem 405(29):9343–9349
Nagarkatti R, Bist V, Sun S, de Araujo FF, Nakhasi HL, Debrabant A (2012) Development of an aptamer-based concentration method for the detection of Trypanosoma cruzi in blood. PLoS One 7(8):e43533
Nonaka Y, Yoshida W, Abe K, Ferri S, Schulze H, Bachmann TT, Ikebukuro K (2012) Affinity improvement of a VEGF aptamer by in silico maturation for a sensitive VEGF-detection system. Anal Chem 85(2):1132–1137
Nutiu R, Li Y (2004) Structure-switching signaling aptamers: transducing molecular recognition into fluorescence signaling. Chem Eur J 10(8):1868–1876
Ospina-Villa JD, Zamorano-Carrillo A, Castañón-Sánchez CA, Ramírez-Moreno E, Marchat LA (2016) Aptamers as a promising approach for the control of parasitic diseases. Braz J Infect Dis 20(6):610–618
Ospina-Villa J, López-Camarillo C, Castañón-Sánchez C, Soto-Sánchez J, Ramírez-Moreno E, Marchat L (2018) Advances on aptamers against protozoan parasites. Genes 9(12):584
Parekh P, Tang Z, Turner PC, Moyer RW, Tan W (2010) Aptamers recognizing glycosylated hemagglutinin expressed on the surface of vaccinia virus-infected cells. Anal Chem 82(20):8642–8649
Park J-W, Lee SJ, Choi E-J, Kim J, Song J-Y, Gu MB (2014) An ultra-sensitive detection of a whole virus using dual aptamers developed by immobilization-free screening. Biosens Bioelectron 51:324–329
Pereira RL, Nascimento IC, Santos AP, Ogusuku IE, Lameu C, Mayer G, Ulrich H (2018) Aptamers: novelty tools for cancer biology. Oncotarget 9(42):26934
Pfeiffer F, Rosenthal M, Siegl J, Ewers J, Mayer G (2017) Customised nucleic acid libraries for enhanced aptamer selection and performance. Curr Opin Biotechnol 48:111–118
Radom F, Jurek PM, Mazurek MP, Otlewski J, Jeleń F (2013) Aptamers: molecules of great potential. Biotechnol Adv 31(8):1260–1274
Ramos E, Piñeiro D, Soto M, Abanades DR, Martín ME, Salinas M, González VM (2007) A DNA aptamer population specifically detects Leishmania infantum H2A antigen. Lab Investig 87(5):409–416
Ramos E, Moreno M, Martín ME, Soto M, Gonzalez VM (2010) In vitro selection of Leishmania infantum H3-binding ssDNA aptamers. Oligonucleotides 20(4):207–213
Rocheteau P, Chatre L, Briand D, Mebarki M, Jouvion G, Bardon J et al (2015) Sepsis induces long-term metabolic and mitochondrial muscle stem cell dysfunction amenable by mesenchymal stem cell therapy. Nat Commun 6:10145
Roh C, Kim SE, Jo S-K (2011) Label free inhibitor screening of hepatitis C virus (HCV) NS5B viral protein using RNA oligonucleotide. Sensors 11(7):6685–6696
Scallan E, Hoekstra RM, Angulo FJ, Tauxe RV, Widdowson M-A, Roy SL, Griffin PM (2011) Foodborne illness acquired in the United States—major pathogens. Emerg Infect Dis 17(1):7–15
Shen H, Wang J, Liu H, Li Z, Jiang F, Wang F-B, Yuan Q (2016) Rapid and selective detection of pathogenic bacteria in bloodstream infections with aptamer-based recognition. ACS Appl Mater Interfaces 8(30):19371–19378
Shum KT, Tanner JA (2008) Differential inhibitory activities and stabilisation of DNA aptamers against the SARS coronavirus helicase. Chembiochem 9(18):3037–3045
Sidhu R, Rong Y, Vanegas D, Claussen J, McLamore E, Gomes C (2016) Impedance biosensor for the rapid detection of Listeria spp. based on aptamer functionalized Pt-interdigitated microelectrodes array. Paper presented at the Smart Biomedical and Physiological Sensor Technology XIII
Song K-M, Cho M, Jo H, Min K, Jeon SH, Kim T, Ban C (2011) Gold nanoparticle-based colorimetric detection of kanamycin using a DNA aptamer. Anal Biochem 415(2):175–181
Song K-M, Lee S, Ban C (2012) Aptamers and their biological applications. Sensors 12(1):612–631
Suh S-K, Song S, Oh H-B, Hwang S-H, Hah SS (2014) Aptamer-based competitive binding assay for one-step quantitation of hepatitis B surface antigen. Analyst 139(17):4310–4314
Sung HJ, Choi S, Lee JW, Ok CY, Bae Y-S, Kim Y-H, Lee W, Heo K, Kim I-H (2014) Inhibition of human neutrophil activity by an RNA aptamer bound to interleukin-8. Biomaterials 35(1):578–589
Sypabekova M, Bekmurzayeva A, Wang R, Li Y, Nogues C, Kanayeva D (2017) Selection, characterization, and application of DNA aptamers for detection of Mycobacterium tuberculosis secreted protein MPT64. Tuberculosis 104:70–78
Tombelli S, Minunni M, Luzi E, Mascini M (2005) Aptamer-based biosensors for the detection of HIV-1 Tat protein. Bioelectrochemistry 67(2):135–141
Torres-Chavolla E, Alocilja EC (2009) Aptasensors for detection of microbial and viral pathogens. Biosens Bioelectron 24(11):3175–3182
Tuerk C, Gold L (1990) Systematic evolution of ligands by exponential enrichment: RNA ligands to bacteriophage T4 DNA polymerase. Science 249(4968):505–510
Ulrich H, Martins AHB, Pesquero JB (2004) RNA and DNA aptamers in cytomics analysis. Cytometry A 59(2):220–231
van den Kieboom CH, van der Beek SL, Mészáros T, Gyurcsányi RE, Ferwerda G, de Jonge MI (2015) Aptasensors for viral diagnostics. TrAC Trends Anal Chem 74:58–67
Wandtke T, Woźniak J, Kopiński P (2015) Aptamers in diagnostics and treatment of viral infections. Viruses 7(2):751–780
Wang RE, Wu H, Niu Y, Cai J (2011) Improving the stability of aptamers by chemical modification. Curr Med Chem 18(27):4126–4138
Wang J, Wu X, Wang C, Shao N, Dong P, Xiao R, Wang S (2015) Magnetically assisted surface-enhanced Raman spectroscopy for the detection of Staphylococcus aureus based on aptamer recognition. ACS Appl Mater Interfaces 7(37):20919–20929
Wongphatcharachai M, Wang P, Enomoto S, Webby RJ, Gramer MR, Amonsin A, Sreevatsan S (2012) Neutralizing DNA aptamers against swine influenza H3N2 viruses. J Clin Microbiol 51(1):46–54
Wu YX, Kwon YJ (2016) Aptamers: the “evolution” of SELEX. Methods 106:21–28
Wu S, Duan N, Shi Z, Fang C, Wang Z (2014) Simultaneous aptasensor for multiplex pathogenic bacteria detection based on multicolor upconversion nanoparticles labels. Anal Chem 86(6):3100–3107
Wu S, Duan N, Qiu Y, Li J, Wang Z (2017) Colorimetric aptasensor for the detection of Salmonella enterica serovar typhimurium using ZnFe2O4-reduced graphene oxide nanostructures as an effective peroxidase mimetics. Int J Food Microbiol 261:42–48
Yamamoto R, Kumar PK (2000) Molecular beacon aptamer fluoresces in the presence of Tat protein of HIV-1. Genes Cells 5(5):389–396
Yang CJ, Jockusch S, Vicens M, Turro NJ, Tan W (2005) Light-switching excimer probes for rapid protein monitoring in complex biological fluids. Proc Natl Acad Sci 102(48):17278–17283
Zelada-Guillén GA, Riu J, Düzgün A, Rius FX (2009) Immediate detection of living bacteria at ultralow concentrations using a carbon nanotube based potentiometric aptasensor. Angew Chem Int Ed 48(40):7334–7337
Zhang H, Ma X, Liu Y, Duan N, Wu S, Wang Z, Xu B (2015) Gold nanoparticles enhanced SERS aptasensor for the simultaneous detection of Salmonella typhimurium and Staphylococcus aureus. Biosens Bioelectron 74:872–877
Zhao W, Chiuman W, Brook MA, Li Y (2007) Simple and rapid colorimetric biosensors based on DNA aptamer and noncrosslinking gold nanoparticle aggregation. Chembiochem 8(7):727–731
Zheng X, Hu B, Gao S, Liu D, Sun M, Jiao B, Wang L (2015) A saxitoxin-binding aptamer with higher affinity and inhibitory activity optimized by rational site-directed mutagenesis and truncation. Toxicon 101:41–47
Acknowledgments
Authors acknowledge to Indian Institute of Technology Roorkee for providing library.
Conflict of Interest
The authors declare no conflict of interest.
Author information
Authors and Affiliations
Editor information
Editors and Affiliations
Rights and permissions
Copyright information
© 2019 Springer Nature Singapore Pte Ltd.
About this chapter
Cite this chapter
Kumar, V., Sharma, A. (2019). Recent Updates for Isolation of Aptamers for Various Biothreat Agents Using Different Strategies and Their Role in Detection Applications. In: Yadav, G., Kumar, V., Aggarwal, N. (eds) Aptamers. Springer, Singapore. https://doi.org/10.1007/978-981-13-8836-1_2
Download citation
DOI: https://doi.org/10.1007/978-981-13-8836-1_2
Published:
Publisher Name: Springer, Singapore
Print ISBN: 978-981-13-8835-4
Online ISBN: 978-981-13-8836-1
eBook Packages: Biomedical and Life SciencesBiomedical and Life Sciences (R0)