Abstract
Silicatein is the first protein isolated from the silicon biominerals and characterized as constituent of the axial filament in the silica spicules of the demosponge Tethya aurantia, by significant sequence similarity with cathepsin L, an animal lysosomal protease, and as a catalyst of silica polycondensation at neutral pH and room temperature. This protein was then identified in a wide range of the class Demospongiae and in some species of the class Hexactinellida. Our attempt to isolate silicatein from the silica skeleton of Euplectella was unsuccessful, but instead we discovered glassin, a protein directing acceleration of silica polycondensation and sharing no significant relationship with any proteins including silicatein. The present study aims to verify the existence of silicatein by exploring the whole genome DNA sequence database of E. curvistellata with the sequence similarity search. Although we identified the sequences of glassin, cathepsin L and chitin synthetase, an enzyme synthesizing chitin, which has already been found in the silicon biominerals in E. aspergillum, silicatein failed to be identified. Our result indicates that silicatein is not essential for poriferan silicon biomineralization in the presence of glassin.
You have full access to this open access chapter, Download conference paper PDF
Similar content being viewed by others
Keywords
1 Introduction
Silicon biominerals are produced by the living organisms through physiological activities in contrast to silicon-based manmade products, often manufactured through processes with high energy consumption and harsh impacts on the environment. Understanding of the mechanisms on silicon biomineralization is expected to offer the prospect of developing environmentally benign routes to synthesize silicon-based materials. Silicon biominerals generally contain a small amount of organic substances, which may help production of silicon biominerals at physiological conditions.
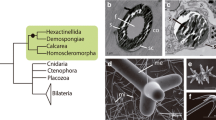
Phylum Porifera (sponges) consists of four classes, Hexactinellida (glass sponges), Demospongiae (demosponges), Calcarea (calcareous sponges), and Homoscleromorpha, among which Hexactinellida, Demospongiae, and Homoscleromorpha produce silica biominerals while calcium carbonate biominerals occur in Calcarea.
The demosponge Tethya aurantia produces a large quantity of silicon biomineral in a form of silica as needlelike structures or spicules, allowing us to isolate and analyze the organic molecules occluded in the biomineral. Silicatein, the first protein isolated from silicon biomineral and characterized, constitutes the axial filament in the spicules; shares significant sequence similarity with cathepsin L, an animal lysosomal protease; and catalyzes silica polycondensation at neutral pH and room temperature (Shimizu et al. 1998; Cha et al. 1999). This protein and its gene were then identified in a wide range of the class Demospongiae (Krasko et al. 2000; Pozzolini et al. 2004; Funayama et al. 2005; Müller et al. 2007). In addition, PCR products encoding a partial silicatein sequences were amplified in the class Hexactinellida including Crateromorpha meyeri (Müller et al. 2008a), Monorhaphis chuni (Müller et al. 2008b), and Euplectella aspergillum (unpublished data. The sequence was deposited to GenBank database by Müller et al. in 2011. The accession number FR748156). Our attempt to isolate silicatein from the silica skeleton of the E. aspergillum and E. curvistellata was unsuccessful, but instead we discovered glassin as a protein directing acceleration of silica polycondensation (Shimizu et al. 2015). Sequences encoding silicatein have not identified from the transcriptome analysis of Aphrocallistes vastus by Riesgo et al. (2015). Veremeichik et al. (2011) tried to isolate silicatein genes from Pheronema raphanus, Aulosaccus schulzei, and Bathydorus levis, resulting in only identification of Aulosaccus sp. silicatein-like sequence with cysteine at the catalytic residue instead of serine as seen in silicateins of other species. Collectively, the existence of silicateins has been unsettled in Hexactinellida.
The present study aims to verify the existence of genetic information on silicatein by exploring the whole genome DNA sequence database of E. curvistellata with the sequence similarity search and discusses on relationship of silicatein and glassin in silicon biomineralization of Porifera.
2 Materials and Methods
Live specimens of E. curvistellata were collected at a depth of 236 m at 32°30 N, 129°10 E in the East China Sea on March 4, 2012, as described previously (Shimizu et al. 2015) and then stored in ethanol at −20 °C. Genomic DNA was extracted from the specimen stored in ethanol with DNeasy Blood & Tissue Kit (QIAGEN, Hilden, Germany). The genomic DNA library was prepared from mechanically fragmented genomic DNA with TruSeq DNA prep kit (Illumina, San Diego, CA, USA). Then, the library was sequenced with MiSeq (Illumina) three times. The raw reads were trimmed and assembled using Genomics Workbench (CLC Bio Inc., Aarhus, Denmark). Sequence similarities were analyzed with NCBI BLAST program.
3 Results and Discussion
3.1 Construction of Whole Genome DNA Library of E. curvistellata
The library of E. curvistellata whole genome DNA was constructed with the next-generation DNA sequencer. Total three runs of sequencing gave rise to 27,939,250 reads, being assembled to 442,583 contigs with the average length of 427 and the median length of 420 (Table 16.1). The longest contig covers 145,960 while the shortest is 18 nucleotides. Total number of the nucleotides reached to 190,209,345. This number can be roughly considered as a genome size of the species, with fairly matching to that of Amphimedon queenslandica, being 167 Mbp (Srivastava et al. 2010).
To evaluate the quality of the library, we run the blast program with Aphrocallistes vastus Cox3 gene (GenBank accession no. EU000309.1) (Rosengarten et al. 2008) as a query. As a result, we obtained the single contig_1075 containing not only Cox3 gene but also the whole mitochondrion DNA sequence, 19,700 bp. This result indicates that the library is qualitatively sound and can be useful for gene searching.
3.2 Search for Silicatein Gene
The axial filament was obtained in the intact form by dissolving the silica spicules of T. aurantia (Shimizu et al. 1998). Although the axial filament was observed in the cross section of Euplectella silica spicules under the scanning electron microscope (Weaver et al. 2007), the axial filaments or any filamentous materials were not obtained in our attempt. The extract contained proteins, but these proteins had no silicatein sequences as long as we examined. On the other hand, a partial silicatein cDNA from E. aspergillum was archived in DNA sequence database (FR748156) (Table 16.2). In addition, silicateins or silicatein-like sequences have been reported from the hexactinellid sponges Aulosaccus sp. (Veremeichik et al. 2011), C. meyeri (Müller et al. 2008a), and M. chuni (Müller et al. 2008b).
To verify the silicatein sequence in E. curvistellata genome, the local blast program was executed with these hexactinellid silicatein sequences as well as T. aurantia silicateins as queries and E. curvistellata genomic DNA library as a database. For the partial silicatein cDNA from E. aspergillum, no contig with E values less than 10 was hit. Similarly, no hit was obtained when T. aurantia silicateins α (AF032117) and β (AF098670), Aulosaccus sp. silicatein-like (ACU86976), C. meyeri silicatein (CAP49202), and M. chuni silicatein (CAZ04880) were used as queries.
The amino acid sequence KNSWG was widely conserved in silicateins and cathepsin L; 296–300 of T. aurantia silicatein α (Shimizu et al. 1998, AF032117), 296–300 of Suberites domuncula silicatein (Krasko et al. 2000, AJ272013), 292–296 of S. domuncula cathepsin L (Müller et al. 2003, AJ784224), and 299–303 human cathepsin L (Gal and Gottesman 1988, X12451). In the case of this sequence used as a query, contig_7,117 (8869 bp) was hit. The contig contains the stop codon, but not the first Met. The 5′ region, contig_50860, was obtained by running blast program with the contig_7,117 as a query. Total length of the coding region is 1343 bp composed of 4 exons coding 324 amino acid residues and 3 introns. The predicted protein sequence is more similar to those of sponge and human cathepsin L than silicateins. In addition, cysteine at the position corresponding to the catalytic residue and the surrounding sequences in cathepsin L are conserved in the contig, indicating that the gene encodes cathepsin L but not silicatein. The boundaries of all the four exons in the cathepsin L of E. curvistellata are identical to those of exon 2/exon 3, exon 3/exon 4, and exon 4/exon 5 in human cathepsin L gene consists of eight exons and seven introns (Chauhan et al. 1993), suggesting that the exon-intron structure of cathepsin L in E. curvistellata is conserved in the human gene.
The result of our blast search for silicatein in E. curvistellata genome is consistent with the fact that the silicatein or silicatein-like proteins were not obtained in dissolution of silica spicules. Previous transcriptome analysis concluded that any silicatein gene was identified in the hexactinellid A. vastus (Riesgo et al. 2015). It is unlikely that silicatein exists in all species of the class Hexactinellida. However, further research should be performed using the hexactinellid species which have been reported to have the evidence for the existence of silicateins before the conclusion is drawn.
3.3 Search of Genes Associated with Silicon Biomineralization
Genes for glassin were assigned by conducting the similarity research with glassin cDNA as a query. The two contigs 14,569 and 22,997 cover 5′ and 3′ regions of glassin gene, respectively, while overlapping each other. Some mismatches were observed in the overlapped and 3′ regions, indicating the assembly in complicated sequences including the repetitive sequences is incomplete. Therefore, further refinement on the library may be required.
Ehrlich and Worch (2007) reported chitin in E. aspergillum as an organic component of their silicious skeletal systems. A gene encoding chitin synthase was searched using A. queenslandica chitin synthase2 and 3-like protein sequences (XP_011402997 and XP_003389565, respectively) as queries, and contig_18,557 (12,682 bp) containing 4335 bp open reading flame encoding 1445 amino acid residues was obtained. Our result suggests that Euplectella is capable of chitin synthesis and thus is consistent with previous observation on occurrence of chitin in Euplectella.
4 Conclusion
The present study aims to verify the existence of genetic information on silicatein by exploring the whole genome DNA sequence database of E. curvistellata with the sequence similarity search. We identified the sequences of glassin, cathepsin L and chitin synthetase, an enzyme synthesizing chitin, which has already been found in the silicon biominerals in E. aspergillum (Ehrlich and Worch 2007). However, silicatein failed to be identified in the genome data consistent with the previous extraction experiment (Shimizu et al. 2015). Although PCR products encoding partial silicatein sequences have been amplified in some hexactinellid sponges (Müller et al. 2008a, b), silicatein was not identified in the transcriptome analysis of A. vastus (Riesgo et al. 2015). Collectively, the existence of silicatein is not evident in Hexactinellid. At least, silicatein is not essential and glassin is responsible for silicon biomineralization in E. curvistellata. Therefore, the evidences imply that there are at least two ways for silicon biomineralization in Porifera in terms of usage of the protein for silica polymerization, silicatein or glassin, and that the selection of either protein depends on the species but not on the taxonomic classes.
References
Cha JN, Shimizu K, Zhou Y, Christiansen SC, Chmelka BF, Stucky GD, Morse DE (1999) Silicatein filaments and subunits from a marine sponge direct the polymerization of silica and silicones in vitro. PNAS 96:361–365
Chauhan SS, Popescu NC, Ray D, Fleischmann R, Gottesman MM, Troen BR (1993) Cloning, genomic organization, and chromosomal localization of human cathepsin L. J Biol Chem 268:1039–1045
Ehrlich H, Worch H (2007) Sponges as natural composites: from biomimetic potential to development of new biomaterials. In: Hajdu E (ed) Porifera research: biodiversity, innovation & sustainability, vol 28. Museu Nacional, Rio de Janeiro, pp 217–223
Funayama N, Nakatsukasa M, Kuraku S, Takechi K, Dohi M, Iwabe N, Miyata T, Agata K (2005) Isolation of Ef silicatein and Ef lectin as molecular markers of sclerocytes and cells involved in innate immunity in the freshwater sponge Ephydatia fluviatilis. Zool Sci 22:1113–1122
Gal S, Gottesman MM (1988) Isolation and sequence of a cDNA for human pro-(cathepsin L). Biochem J 253:303–306
Krasko A, Lorenz B, Batel R, Schröder HC, Müller IM, Müller WEG (2000) Expression of silicatein and collagen genes in the marine sponge Suberites domuncula is controlled by silicate and myotrophin. Eur J Biochem 267:4878–4887
Müller WEG, Krasko A, Le Pennec G, Schröder HC (2003) Biochemistry and cell biology of silica formation in sponges. Microsc Res Tech 62:368–377
Müller WEG, Schloßmacher U, Eckert C, Krasko A, Boreiko A, Ushijima H, Wolf SE, Tremel W, Müller IM, Schröder HC (2007) Analysis of the axial filament in spicules of the demosponge Geodia cydonium: different silicatein composition in microscleres (asters) and megascleres (oxeas and triaenes). Eur J Cell Biol 86:473–487
Müller WEG, Wang X, Kropf K, Boreiko A, Schloßmacher U, Brandt D, Schröder HC, Wiens M (2008a) Silicatein expression in the hexactinellid Crateromorpha meyeri: the lead marker gene restricted to siliceous sponges. Cell Tissue Res 333:339–351
Müller WEG, Boreiko A, Schloßmacher U, Wang X, Eckert C, Kropf K, Li J, Schröder HC (2008b) Identification of a silicatein (-related) protease in the giant spicules of the deep-sea hexactinellid Monorhaphis chuni. J Exp Biol 211:300–309
Pozzolini M, Sturla L, Cerrano C, Bavestrello G, Camardella L, Parodi AM, Raheli F, Benatti U, Müller WEG, Giovine M (2004) Molecular cloning of silicatein gene from marine sponge Petrosia ficiformis (Porifera, Demospongiae) and development of primmorphs as a model for biosilicification studies. Mar Biotechnol 6:594–603
Riesgo A, Maldonado M, López-Legentil S, Giribet G (2015) A proposal for the evolution of Cathepsin and Silicatein in sponges. J Mol Evol 80:278–291
Rosengarten RD, Sperling EA, Moreno MA, Leys SP, Dellaporta SL (2008) The mitochondrial genome of the hexactinellid sponge Aphrocallistes vastus: evidence for programmed translational frameshifting. J BMC Genomics 9(33)
Shimizu K, Cha J, Stucky GD, Morse DE (1998) Silicatein alpha: Cathepsin L-like protein in sponge biosilica. Proc Natl Acad Sci U S A 95:6234–6238
Shimizu K, Amano T, Bari Md R, Weaver JC, Arima J, Mori N (2015) Glassin, a histidine-rich protein from the silicious skeletal system of the marine sponge Euplectella directs silica polycondensation. Proc Nat Acad Sci USA 112:11449–11454
Srivastava M, Simakov O, Chapman J, Fahey B, Gauthier MEA, Mitros T, Richards GS, Conaco C, Dacre M, Hellsten U et al (2010) The Amphimedon queenslandica genome and the evolution of animal complexity. Nature 466:720–726
Veremeichik GN, Shkryl YN, Bulgakov VP, Shedko SV, Kozhemyako VB, Kovalchuk SN, Krasokhin VB, Zhuravlev YN, Kulchin YN (2011) Occurrence of a silicatein gene in glass sponges (Hexactinellida: Porifera). Mar Biotechnol 13:810–819
Weaver JC, Aizenberg J, Fantner GE, Kisailus D, Woesz A, Allen P, Fields K, Porter MJ, Zok FW, Hansma PK, Fratzl P, Morse DE (2007) Hierarchical assembly of the siliceous skeletal lattice of the hexactinellid sponge Euplectella aspergillum. J Struct Biol 158:93–106
Acknowledgment
This work is supported by JSPS Kakenhi grant number 15 K06581.
Author information
Authors and Affiliations
Corresponding author
Editor information
Editors and Affiliations
Rights and permissions
Open Access This chapter is licensed under the terms of the Creative Commons Attribution 4.0 International License (http://creativecommons.org/licenses/by/4.0/), which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license and indicate if changes were made.
The images or other third party material in this chapter are included in the chapter's Creative Commons license, unless indicated otherwise in a credit line to the material. If material is not included in the chapter's Creative Commons license and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder.
Copyright information
© 2018 The Author(s)
About this paper
Cite this paper
Shimizu, K., Kobayashi, H., Nishi, M., Tsukahara, M., Bito, T., Arima, J. (2018). Exploration of Genes Associated with Sponge Silicon Biomineralization in the Whole Genome Sequence of the Hexactinellid Euplectella curvistellata . In: Endo, K., Kogure, T., Nagasawa, H. (eds) Biomineralization. Springer, Singapore. https://doi.org/10.1007/978-981-13-1002-7_16
Download citation
DOI: https://doi.org/10.1007/978-981-13-1002-7_16
Published:
Publisher Name: Springer, Singapore
Print ISBN: 978-981-13-1001-0
Online ISBN: 978-981-13-1002-7
eBook Packages: Biomedical and Life SciencesBiomedical and Life Sciences (R0)