Abstract
“Why do parasites harm their host?” is a recurrent question in evolutionary biology and ecology, and has several implications for the biomedical sciences, particularly public health and epidemiology. Contrasting the meaning(s) of the concept of “virulence” in molecular pathology and evolutionary ecology, we review different explanations proposed as to why, and under what conditions, parasites cause harm to their host: whereas the former uses molecular techniques and concepts to explain changes and the nature of virulence seen as a categorical trait, the latter conceptualizes virulence as a phenotypic quantitative trait (usually related to a reduction in the host’s fitness). After describing the biology of emerging influenza viruses we illustrate how the ecological and the molecular approaches provide distinct (but incomplete) explanations of the 1918–19 influenza pandemic. We suggest that an evolutionary approach is necessary to understand the dynamics of disease transmission but that a broader understanding of virulence will ultimately benefit from articulating and integrating the ecological dynamics with cellular mechanisms of virulence. Both ecological and functional perspectives on host-pathogens’ interactions are required to answer the opening question but also to devise appropriate health-care measures in order to prevent (and predict?) future influenza pandemics and other emerging threats. Finally, the difficult co-existence of distinct explanatory frameworks reflects the fact that scientists can work on a same problem using various methodologies but it also highlights the enduring tension between two scientific styles of practice in biomedicine.
You have full access to this open access chapter, Download chapter PDF
Similar content being viewed by others
Keywords
These keywords were added by machine and not by the authors. This process is experimental and the keywords may be updated as the learning algorithm improves.
Introduction
The question “why do parasites harm their hosts?” is recurrent in evolutionary biology and ecology, and has several implications for the medical sciences, particularly public health and epidemiology.Footnote 1 The question is perplexing because of its paradoxical aspect. Indeed, one wonders why natural selection favours high virulence if this inevitably results in both the host and the pathogen’s deaths. Shouldn’t host and pathogensFootnote 2 peacefully coevolve, and thus maximize both their chances of survival, instead of engaging in a near-infinite arms race? Very much along these lines, The Lives of a Cell (1974) by American physician Lewis Thomas reflected the conviction that “there is nothing to be gained, in an evolutionary sense, by the capacity to cause illness or death” (Thomas 1974, 77). Thomas’ views on the nature of disease were once widely accepted among medical scientists during the past century.Footnote 3 The possibility of eradicating diseases like smallpox, combined with the belief that evolution was going to naturally wipe out infections, worked together in supporting the idea of the end of infectious diseases (Levins 1994). Physician and epidemiologist Aidan Cockburn, for instance, stated confidently: “it seems reasonable to anticipate that within some measurable time, such as 100 years, all the major infections will have disappeared” (Cockburn 1963, 150). Following the improved control over infections provided by vaccines, antibiotics, and chemotherapy, biomedical authorities in the 1950s and 1960s, particularly in the U.S., ceased to regard infectious diseases as one of the major causes of death and morbidity, and argued, furthermore, that fundamental research on microorganisms could be halted altogether (Burnet 1953 in Fantini 1993). This perspective was also reflected at the political and economic levels. After the “war” on cancer and cardio-vascular diseases was declared in the early 1970s, for instance, the budget of the National Institute of Health (NIH) doubled in 5 years, while the funding for the National Institute of Allergy and Infectious Diseases (NIAID) grew by only 20 % (Krause 1998, 3). The belief in the power of medical technology to conquer infectious diseases with newly developed drugs resulted in the idea that given sufficient time most of these diseases would naturally decline as a result of the evolutionary dynamics that govern host and pathogens’ relation and lead to lower levels of virulence over time (Méthot 2012a; Snowden 2008).
The return of infectious diseases from the early 1980s onwards turned this perspective on its head, however, as the responses of modern medicine seemed no longer adequate in the face of the steep rise of nosocomial infections and the evolution of drug resistance worldwide. Particularly, the acute sense of control over infectious disease felt by many was thrown into disarray with the onset of the HIV pandemic and other emerging infections such as Ebola fever, SARS, and more recently with the return of H1N1 influenza. Partly because “many people find it difficult to accommodate the reality that Nature is far from benign” (Lederberg 1993, 3), the rationale of the “conventional wisdom” (as named by May and Anderson 1983) – namely that hosts and pathogens should coevolve towards a state of harmlessness – was promoted far into the second half of the twentieth century (see Ewald 1994 for a review). An additional reason for the success of this avirulence hypothesis, besides its intuitive soundness, was the fact that no serious alternatives to it were introduced before the late 1970s (Alizon et al. 2009), even though some like zoologist Gordon Ball (1943) did raise important objections to the conventional wisdom. The thesis of a natural decline in the virulence of infectious disease postulated by earlier evolution-led models has been challenged on both theoretical and experimental grounds in the last 30 years. Empirical evidence and advances in modelling in evolutionary ecology (e.g. the trade-off model) have shown, for instance, that the evolution of hosts and parasites into a commensal state is not the vanishing, obligate point it was once held to be, but is rather only one of the possible evolutionary outcomes (Anderson and May 1982; Levin and Pimentel 1981; Ewald 1983; reviewed in Alizon et al. 2009). As biologist Carl Bergstrom recently stressed: “we cannot count on evolution to do our work for us” (Bergstrom 2008, 261). Selective pressures, on the contrary, can drive the emergence of new diseases (Antia et al. 2003). And as some have argued, humans may well be the “world’s greatest evolutionary force” (Palumbi 2001) behind the increased virulence of pathogens.
Through new social and cultural practices we open-up new routes for “viral traffic” (e.g. blood transfusions, organ transplants), foster behavioural changes facilitating pathogens’ transmission (e.g. air travel, migrations, sexual practices, use of drugs, etc.), and introduce “new” pathogens from different parts of the world into immunologically naive populations (Morse 1991, 1993, 1995). This of course adds up to the continuing emergence of human pathogens through zoonotic reservoirs (Wolfe et al. 2007). Infectious diseases continue to be a serious threat to human health, and some diseases once believed to be eradicated might return. Between 1940 and 2004, 335 diseases have emerged in human populations, the majority of them appearing during the 1980s after rapid increase in drug resistance was detected (Jones et al. 2008). Despite the recent steep rise in chronic and degenerative illnesses, emerging infections are still a global challenge for twenty-first century biomedicine and they continue to claim 15 million lives annually (Morens et al. 2004; Fauci 2000). Following the resurgence of infectious diseases as a leading cause of death and morbidity, and the detection of previously unknown disease-causing entities, the idea that newly emerged pathogens have thrown the natural world “out of balance” (Garrett 1994) has garnered a significant amount of scientific attention and has led to the adoption of new international health regulations in order to monitor, limit, and control the spread of communicable diseases (Castillo-Salgado 2010). Here, we explore how, and in what contexts (molecular, ecological, and evolutionary), knowledge claims about disease emergence and changes in virulence are made and justified in the case one specific example: the 1918–19 influenza pandemic.
Emerging diseases are usually defined as diseases whose incidence has significantly increased within a population over a definite period of time (Morse 1995).Footnote 4 As Weir and Mykhalovski (2010) recently observed, two of the most influential books on emerging diseases in the early 1990s (Lederberg et al. 1992; Morse 1993) have stressed the need to investigate factors driving disease emergence from both an ecological and a molecular-genetic point of view. Both books argued that the biology of the host and the pathogen, in addition to their complex interactions in changing ecological and evolutionary contexts, must be carefully considered in order to devise appropriate public health measures. In practice, though, it remains a challenging task to integrate those perspectives. Indeed, our starting point is the current gap – and lack of integration – in the literature between studies of virulence as applied to emerging disease in the biomedical sciences broadly understood and in molecular pathology and evolutionary ecology in particular. Integration is a multi-faceted concept that is often promoted as a promising goal of scientific practice. As discussed by philosophers of science, integration in science is a complex process that encompasses several activities such as methodological integration, data integration, and explanatory integration (O’Malley and Soyer 2012; see also Mitchell 2008), among others.Footnote 5 More rarely is the possibility that integration will fail discussed, however (see O’Malley (2013) for an example of such). As this chapter exemplifies, ecological and molecular methodologies have yet to come together to provide a broader picture of changes in virulence in emerging diseases. Here, we focus particularly on experimental and modelling practices in molecular biomedicine and evolutionary ecology and on their respective explanatory limitations. Very often, explanations of the virulence of a pandemics are constructed as an alternative between knowing the biological nature of the pathogen or that of the environmental conditions that facilitate its transmission. While both consider the nature of the host as part of the disease process, most of the time one branch of the alternative alone is considered as the right (or at least sufficient) explanation while attention to other explanatory schemes is scant. Using the 1918–19 influenza pandemic as a case study of a particularly virulent emerging disease, we illustrate the enduring persistence of two distinct scientific styles of practice in the recent history of virulence studies.
Beginning with a discussion of the evolution of virulence as seen through the lens of ecological and molecular perspectives in biology, we show how each of them conceptualizes both the nature of virulence and emergence in quite different ways. Next, we describe the biology of influenza viruses with a focus on the 1918–19 pandemics and we move on to the ecological-evolutionary explanations of its exceptional virulence, paying attention to the trade-off model, before turning to molecular pathology. We argue that an evolutionary approach is necessary to understand the dynamics of disease transmission and evolution but that a broader understanding of virulence will ultimately benefit from articulating the ecological dynamics with cellular mechanisms of virulence. In sum, both ecological and functional perspectives on host-pathogens’ interactions are required to answer the opening question of this essay but also to devise appropriate health-care measures in order to prevent (and predict?) future influenza pandemics and other emerging threats. The difficult co-existence of distinct explanatory frameworks reflects the fact that scientists can work on a same problem using distinct methodologies (Godfrey-Smith 2006), but it also highlights the enduring tension between two scientific styles of practice in biomedicine.
Functional and Ecological Perspectives on Emerging Diseases and Virulence
Evolutionary biologist Ernst Mayr has long suggested that functional (proximate) and evolutionary (ultimate) perspectives in biology lack unification (Mayr 1961; see Morange 2005). More recently, evolutionary ecologists have argued in the direction of a better integration of those perspectives (Frank and Schmid-Hempel 2008). While Mayr’s point that proximate and ultimate explanations are not alternatives is sound, developmental biology advocates, among others, have persuasively argued that evolutionary questions are relevant to understanding developmental processes, and vice-versa (see Laland et al. 2011 for a review). Today, another, and perhaps equally significant divide, seems to be that between ecological and functional (or proximate) approaches to biological systems and their evolution. As we show, what we call exogenous and endogenous approaches to virulence both make knowledge claims based (sometimes loosely) on evolutionary theory, although each of them invokes one particular aspect of the theory.Footnote 6 Whereas the ecological (or exogenous) style focuses on processes (e.g. selective pressures, population density, within and between host competition, and so on) acting on the hosts and the pathogen, the molecular (or endogenous) style traces the evolutionary pathway, or patterns, of the influenza virus from animal(s) to man, and, by constructing molecular phylogenies, identifies particular genes for pathogenesis and mutation sites within lineages.Footnote 7 In other words, the former analyses one of the main mechanisms of evolution (i.e. natural selection) and the latter describe the path of evolution (i.e. they construct phylogenies) (Ruse 1992). The construction of molecular phylogenetic trees by molecular pathologists reflects the recent “data-driven” trend itself supported by genomics, molecular biology, and the development of high throughput technologies. The use of “evolution” by molecular pathologists is, however, secondary to finding molecular mechanisms for pathogenesis and thus explaining changes in virulence mechanistically.
Each perspective also provides a different way of thinking about disease emergence. Briefly, the endogenous view describes how bacteria and viruses can be transformed into pathogenic, emerging diseases by gaining intracellular and genetic material such as, for instance, a polysaccharide capsule, a large plasmid, a set of virulence genes, or pathogenicity islands (Friesen et al. 2006). These and similar findings have led some to claim that pathogens can evolve in “quantum leap” (Groisman and Ochman 1996). Point mutations allowing the virus to bind to a host receptor also belong to this category. While the capacity to cause disease due to new sets of genes is a crucial aspect of how organisms become pathogenic, this capacity can also occasionally result from genomic deletion and gene loss (Maurelli 2006). In sum, acquisition of novel “virulence factors” (or deletion of other genetic elements) can rapidly lead to the emergence of new diseases or enhanced virulence in some pathogens. For molecular pathologists the concept of virulence is similar to the traditional definition of plant pathologists, i.e. the infectivity: a strain is virulent if it is able to infect a host.Footnote 8 This definition could be traced back to the work of Pasteur, for whom “virulent cultures killed, attenuated ones did not” (Mendelsohn 2002, 3–26, p. 5). A more classical definition is the ability to generate symptoms. In both cases, virulence is an all or nothing trait; it is qualitative and not quantitative. Note that these definitions have the advantage that they can be translated at different levels, for instance at the cellular level, where virulence can be the ability to infect cells.
The ecological or exogenous style adopts another approach to disease emergence, virulence, and evolution. Often described as a two-step process, disease emergence requires the introduction of a pathogen within a population followed by its successful dissemination (Morse 1991, 392–3). The “rules of viral traffic” (Morse 1991) dictate that both steps usually result from one or several changes in the environment, not from a modification in the biological characteristics of the pathogen. For instance, in 1976 a change in the air conditioning system in a hotel in Philadelphia facilitated the spread of Legionellosis, a bacterium usually commensal to humans, which caused an outbreak of fever and pneumonia now known as Legionnaire’s disease. However, there are cases supporting a biological explanation of emergence, for instance when a maladapted strain mutates into a well-adapted strain before going extinct (Antia et al. 2003). The trade-off model developed by Robert May and Roy Anderson, and independently by Paul Ewald, in the early 1980s currently underpins the bulk of the theoretical research on host-pathogen’s interactions in evolutionary ecology.Footnote 9 Put simply, the model postulates the existence of ecological trade-offs between a number of epidemiological variables. As a consequence, the evolution of virulence becomes linked to several factors: host resistance and recovery rate, pathogen transmission rate, the timing of infection life-history events and population density, among others. The trade-off model permits the investigation of the role of environmental changes broadly conceived (including within and between hosts selection) and selective pressures acting on pathogen transmission, and thus on the level of virulence (Alizon 2014).
While molecular geneticists quickly adopted the concepts of virulence genes and pathogenicity islands, evolutionary ecologists working with the trade-off model continued to regard them with suspicion (see Poulin and Combes 1999).Footnote 10 We think this suspicion is probably due to the way virulence is defined. For evolutionary biologists, virulence typically is a quantitative trait that can be measured. Therefore, genes that are sufficient to render a pathogen virulent and essentially act as a qualitative trait are difficult to fit into the picture. Furthermore, there is no such thing as pathogen virulence alone in ecology. Virulence, typically, is a “shared trait” that results from the interaction between a host genotype, a parasite genotype and their environment. In other words, some parasite genotypes might only cause virulence when they infect some host genotypes or some parasites may only be virulent to hosts in certain contexts (e.g. starvation). For evolutionary biologists and ecologists, virulence is the harm a pathogen does to its host, i.e. the reduction in host fitness due to the infection (Read 1994). Fitness is notoriously difficult to evaluate but arguably the two most common measures are lifespan and fecundity.Footnote 11 One problem is that a pathogen strain described as being very virulent in vitro could turn out to be mild in vivo (and vice-versa). Furthermore, recent work shows that levels of virulence can actually be the result of the immune system’s over-response itself (see Graham et al. 2005 for a review). In the end, evolutionary ecologists focus on a combination of within-host processes when they refer to virulence. Importantly, this does not mean that they disregard the molecular processes that lead to virulence. For instance, studies have shown that immune-pathology contributions to virulence lead to a different evolutionary outcome than “virulence factors” produced by pathogens (Alizon and van Baalen 2005).
Recent explanations advanced to account for the rapid changes in virulence during the 1918–19 influenza pandemic reflect the polarity between ecological and molecular explicative strategies. Applying the trade-off model to the 1918–19 pandemic, Paul Ewald has argued that the proximity of soldiers in the trenches, the hospitals, the transport, and the military camps during World War I greatly facilitated transmission of the virus from host to host. High viral replication rate by natural selection was therefore favoured, which resulted in exceptionally high virulence and the high level of mortality of the pandemic (Ewald 1991, 1994, 1996). But since the late 1990s, molecular pathology has provided an alternative viewpoint on the evolution of virulence in the pandemic. The identification of the viral RNA from frozen bodies and wax blocks in the U.S. and its further sequencing has led to a renewed emphasis on genetic and molecular determinants of the virus as being the most important cause of this dramatic event (see Holmes 2004). According to molecular pathologist Jeffrey Taubenberger, one of the leading scientists involved in reviving the 1918 influenza strain, “it is possible that a mutation or reassortment occurred in the late summer of 1918, resulting in significantly enhanced virulence” (2005, 90). Taubenberger believes that this “unique feature” of the 1918 virus – its extreme virulence – “could be revealed in its [genetic] sequence” (2005, 90). Both approaches – the exogenous and the endogenous – evolved along parallel lines during most of the twentieth century, and though the concept of emerging infectious diseases brought them closer to one another in the 1990s, we show how they remain in tension (Méthot 2012b). Before describing in more details the potentials and limits of these two perspectives we first describe significant aspects of the biology of influenza viruses.
The Biology of Influenza Viruses and the 1918–19 Pandemic
The natural history and ecology of influenza A virus has been extensively studied (Webster 1999, 1993; Webster et al. 1992; Webster and Rott 1987). The virus’ natural reservoir is the wild waterfowl, as supported by the fact that species of wild duck are not affected by the virus and remain “healthy”. The virus replicates inside the host, mostly in the intestinal tract, and is then washed into the ponds where ducks live and breed (Webster 1993). The relative harmlessness of this relationship is similar to the way myxoma virus is adapted to its natural host, the South American rabbit (see Fenner and Fantini 1999). The family tree of influenza viruses contains two genera: one that includes influenza A and B viruses and the other influenza C viruses. The two genera are distinct in terms of host range and virulence factors. Type A is the most common of all, and can infect a wide range of hosts, including, pigs, horses, seals, whales and birds. This type of virus is also the most redoubtable as it has the potential to cause pandemics. Type B is believed to infect only humans (especially young children) and Type C (another genus) can infect both humans and swine. In this sense influenza can hardly be regarded as a “single disease” (Johnson 2006, 10).
Influenza viruses are enveloped negative strand RNA viruses and belong to the genus Orthomyxoviridae (Taubenberger 2005). The virus of the Spanish flu pandemic belongs to the type A influenza, known as H1N1. Influenza A and B viruses contain eight discrete gene segments, coding for at least one protein. The surface of influenza A viruses is covered by three types of proteins hemmagglutinin (HA), neuraminidases (NA) and matrix 2 (M2). The structural configuration of HA proteins is that of a triangular spike. These spikes allow the virus to bind to red blood cells by causing the latter to agglutinate (i.e. hemmagglutinin). They facilitate entrance into the host and they trigger the infective processes. Once the infection is over, antibodies responding to hemmagglutinin spikes are formed, allowing the immune system to recognize the signature of the viral strain in case of another infection episode. In contrast, neuraminidases (NA) also form spikes on the surface coat of the virus but their function is to cleave glycoproteins into two and to facilitate the propagation of the virus from cell to cell. NA proteins open-up cells for infection, so to speak. Antiviral drugs target NA in order to block their exit, and antibodies to NA are also produced after the infection. Influenza A viruses are further subdivided into serological types, which is the genetic characterization of the surface glycoproteins HA and NA. 16 HA and 9 NA proteins have been described to date. These surface glycoproteins define the virus’ identity in terms of what the immune system detects and attacks. The different major families of flu are combinations of the two, hence the designation “H5N1” for the recent threat. The 1918 virus was H1N1.
The genes coding for these glycoproteins can reassort (i.e. reshuffle) due to two processes known as antigenic drift and antigenic shift. The former consists in the accumulation of point mutations in the genome of the virus, modifying both the shape and the electric charge of viral surface antigens and preventing their recognition by the antibodies of the host that were developed in reaction to previous exposures to the virus. The need to update the influenza vaccines every year illustrates the evolutionary success of antigenic drift. In contrast, antigenic shifts refer to the introduction of whole or part of influenza genes into viruses that circulate among human populations. This form of genetic reassortment or reshuffling occurs especially in swine that act as “mixing vessels” for the viral strains and are considered the intermediate host between birds and humans (Webster and Kawaoka 1994). The introduction of a new hemmagglutinin gene (HA) is often hailed as the responsible factor for increased virulence (Bush 2007). The fast reassortment of nucleotides and the high rate of mutation in influenza viruses result in influenza posing a continual threat for human and animal health. As a result, influenza is regarded as being a continually “re-emerging” disease (Webby and Webster 2003; Webster and Kawaoka 1994), and international efforts are made to understand why the 1918–19 pandemic was so exceptionally virulent. The motivation behind these global efforts in gaining a better understanding of this pandemic is to draw lessons from the past in order to be better prepared for the rise of future influenza and other viral pandemics.
Recorded history suggests that the first influenza pandemic occurred in 1580. Beginning in Asia, it rapidly spread to Africa, America and to Europe. Between the eighteenth and the nineteenth century, medical historians identified (at least) 8 pandemics out of 25 epidemics of influenza A virus (Beveridge 1992). The most devastating pandemic, however, occurred in 1918–19 (Fig. 1).Footnote 12 The emergence of the (misnamed) “Spanish” influenza pandemic of 1918–19 is the first of the three major influenza pandemics that occurred during the past century – and is regarded as one of the most devastating episodes in medical history (McNeill 1976).Footnote 13 Once described as “the biggest unsolved problem of theoretical epidemiology and public health practice” (Burnet and Clark 1942), its consequences rendered many wary about the emergence of respiratory disease pandemics in a near future (Webby and Webster 2003). In addition to the 1918–19 pandemic, two other major influenza pandemics occurred in 1957–58 (“Asian” influenza, H2N2) and in 1968 (“Hong Kong” influenza, H3N2). The death of David Lewis, a soldier at the military camp of Fort Dix in the U.S., and the infection of a few hundreds of others in 1976 led public health authorities to believe they were facing a new influenza epidemic. Amidst some scepticism, vaccines against H1N1 flu were quickly stockpiled as President Ford gave the green light to mass vaccination. However, no epidemic occurred while a number of vaccinated individuals came down with Guillain-Barré syndrome, an autoimmune disease, a few weeks later (see Krause 1998). One year later, in 1977, the H1N1 virus, which had disappeared in 1957, reappeared (the so-called “Russian” influenza) in the Soviet Union and spread to Taiwan, the Philippines, Singapore, and within 10 months had reached South America and New Zealand. The virus was similar to a virus isolated in the U.S. in 1950 at Fort Warren and had perhaps been accidentally released from a laboratory located in the former Soviet Union (Berche 2012, 127). Affecting mostly individuals born after 1957, this virus coexisted with the H3N2 virus until 2009, when a new variant of H1N1 emerged (the “Swine” flu, which is the latest pandemic) that replaced the 1977 variant. In comparison to the Spanish flu pandemic of 1918, the Hong Kong and the Asian pandemics were more “benign”, the former causing between 1.5 and 2 million deaths, and the latter 1 million. The recent H5N1 pandemic caused a few deaths only between 1997 and 2004 (Taubenberger 2005, 87). Despite the (crucial) facts that antibiotics were available during the second two pandemics, and that medical care had significantly improved and was more efficient after 1950, this raises the question: why was the 1918–19 pandemic so deadly to humans?
Three Significant Aspects of the 1918–19 Pandemic
A Western Origin
A first important aspect of the 1918–19 influenza pandemic is its likely Western origin. In part because of its extensive pig-duck farming industry, China was previously singled out as the possible origin of most influenza pandemics. However, whereas most pandemics to have befallen man have come from China (Morse 1993, 17) the “Spanish” flu originated (likely) from France as early as 1916 causing acute respiratory symptoms closely resembling the phenotype of the disease during the 1918–19 flu pandemic (Oxford et al. 2002, 2005). Some have recently argued that there was an early wave of influenza in New York between February and April 1918 (Olson et al. 2005). The precise geographical origins of the 1918 pandemic are still a matter of debate, however.Footnote 14 The world’s deadliest flu pandemic kicked off in October 1918 and in just a few months, the virus killed between 30 and 40 million people (Philips and Killingray 2003; Johnson and Mueller 2002; Crosby 1989; some estimate deaths to number about 50 million, see McNeil 1976). According to the “three waves theory”, influenza swept through all five continents in three recurrences. The first wave (or the “spring wave”) of the flu started in March in the U.S. (Mid West) before moving to Europe, then to Asia and North Africa before reaching Australia in July 1918. While morbidity was high, mortality was not higher than the habitual norm (Reid et al. 2001, 81). The second wave (or “fall wave”), however, was highly devastating and rapidly went extinct after causing millions of deaths worldwide, with peaks in October and November. It started in late August 1918 and within 1 week reports of the virus came from distant cities, including Boston (U.S.), Freetown (Africa), and Brest (France). On many accounts, this second wave lasted until November. The speed at which the virus circulated makes it difficult to pinpoint one specific location as being “the” source of the pandemic but a Western origin appears to be the most plausible hypothesis according to the available evidence. Reports indicated a further third wave that hit in the first months of 1919 but was much less severe (Burnet and Clark 1942; Barry 2004a). However, the three waves pattern of the pandemic is not uniformly applicable to all countries; for instance Australia experienced a single occurrence of the flu pandemic (Johnson and Mueller 2002; Morens and Taubenberger 2009).
Signs, Symptoms, and the Age Group of the Victims
In 1918, during an attack of influenza most victims died of secondary infections as death often resulted from bacteria invading the lungs of immunocompromised individuals (Burnet and Clark 1942). Symptoms lasted generally between 2 and 4 days and could, more rarely, be extended up to 2 weeks. The respiratory disease was characterized by fever, body pain, and severe headaches. Without the possibility of treating patients with antibiotics, bacteria turned “those vital organs [lungs] into sacks of fluids […] effectively drowning the patient” (Philips and Killingray 2003, 5). People therefore died within just a few days of hemorrhagic pulmonary oedema and other lung afflictions (Bush 2007; see also Taubenberger et al. 2000). Related to this, the second striking aspect of the 1918 influenza pandemic is the young age of the victims, which was qualitatively distinctive: most of them were men, supposedly healthy, of between 20–40 years old (some say 25–35), irrespective of whether the country was involved in the war or not. Instead of forming a U shaped mortality curve, the shape of the 1918 pandemic was W shaped. An additional peak (the central peak in the W) represents the male victims of the flu. Figure 2 (above) shows the U shaped curve of 1917 and the W shaped one of 1918. The distribution of deaths on this curve reflects the virulence of the pandemic and underlines the pattern of mortality of a group usually not affected by seasonal flu.
Age distribution of death rates from influenza and pneumonia in the United States death registration area, 1917 and 1918. Death rate is deaths per 100,000 person-years lived. Data from US Department of Health, Education, and Welfare 1956. In Noymer (2010, 141)
Exceptional Virulence
Finally, the third and most significant feature of the 1918 pandemic was its lethality: the disease was of exceptional virulence and estimates suggest that the pandemic claimed more victims than the First World War (McNeil 1976). This central aspect was almost universally recognized as being somewhat unusual and very specific to it although the estimates of fatalities during the twentieth century vary between 20 and 50 million deaths (Johnson and Mueller 2002).
Influenza type A viruses are not at all uncommon, and strains had circulated in human populations for a few centuries before since a few centuries when the 1918 pandemic broke out. In the United States alone only, annual death tolls related to seasonal influenza are estimated to be about 30,000 individuals (Thompson et al. 2003). Seasonal outbreaks of influenza normally last a few weeks and then disappear abruptly; they result from influenza viruses present in human populations that are able to infect individuals due to antigenic drift. On occasions, however, the virus can infect up to 40 % of the world population. During these pandemic years, in contrast, the number of deaths rises way above the average, claiming millions of victims all around the globe. In the course of seasonal epidemics strains of influenza type A and B can sometimes coexist, if at different frequencies among populations. So why was the Spanish influenza so devastating? Recent work in molecular biology argues that the waves pattern, the group mortality, and the clinical course of the disease “may find their explanation in genetic features of the 1918 virus” (Reid et al. 2001, 86). Others, however, defend the view that the changes in virulence result from significant changes in the wider ecological context in which the outbreak occurred (Ewald 1994). In the next sections, we review both ecological-environmental and molecular-led approaches to this problem, we indicate some of the limitations of each and we suggest that a better integration of those perspectives would lead to positive outcomes regarding prediction, prevention, and preparedness in the face of other similar influenza and other bacterial or viral pandemics.
Evolutionary Epidemiology and Environmental Explanations of Disease Emergence
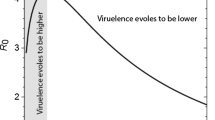
From an ecological point of view, for a disease to emerge in a population the basic reproductive rate of the pathogen (R 0 ) must be higher than 1, where R 0 is the average number of secondary infections that follow from one infected individual in a wholly susceptible population (Anderson and May 1991). In other words, a pathogen must cause at least one subsequent infection to persist in the host population. The classical formula used to capture the trade-off model is as follows:
where R 0 serves as a measure of Darwinian fitness of the pathogen at the epidemiological level. In the denominator are α, the host mortality due to the infection (i.e. the virulence), μ, the rate of microparasite independent-mortality and γ is the rate of recovery of the host. The inverse of μ + α + γ is the average duration of the infection. In the numerator, we have β, the transmission rate, and S, the host population size. Importantly, one should not confuse β, which is a rate (number of infections per unit of time and per susceptible host in the population) and R 0 , which is roughly the number of new hosts infected over the whole duration of the infection. Overall, this expression indicates that parasite fitness is the product between the number of secondary infections generated per unit of time and the duration of the infection. Any animal that produces less than one offspring over its lifetime infections generating less than 1 secondary infection will eventually become extinct and die out.
This R 0 > 1 threshold is of course a simplification. For instance, in the early stages of an outbreak, emerging pathogens infect very few hosts which means that they are particularly prone to extinction going extinct due to stochastic effects. In fact, it can be shown that in an ideal situation where all the hosts would be identical, the probability of emergence of a pathogen with an R 0 strictly greater than unity is only of 1-1/R 0 , due to these stochastic effects (Diekmann and Heesterbeek 2000). Conversely, pathogens with R 0 < 1 can nevertheless be dangerous because they can persist in the population for a while stochastically, which leaves time for a variant with an R 0 > 1 to evolve (Antia et al. 2003). In other words, the transmission between hosts (in these cases humans) must be effective for disease emergence to occur. Historically, the first example of a trade-off came from the analysis of myxoma virus infecting rabbits (Anderson and May 1982; Fenner and Fantini 1999). However, since then, clearer examples have been worked out. Fraser et al. (2007), for instance, combined data on virulence from an Amsterdam cohort and data on transmission rate in discordant HIV-infected couples from a Rakai cohort to show that individuals with a higher set-point viral load (i.e. the viral load during the asymptomatic stage, which has the property to often remain constant over several years) have a shorter lifespan and a higher transmission rate. When they combined host lifespan and virulence together to obtain a measure of parasite fitness (i.e. R 0 ), they found that viruses with an intermediate virus load achieved the highest fitness. They also show the observed abundance of virus loads in a human population follows the distribution virus fitnesses.
The classic trade-off model focuses primarily on pathogen populations and their evolution. It often ignores host evolution because generation times for hosts (here, humans) tend to be much longer and so evolution in the host population is likely to be slow. From a trade-off model perspective, pathogens’ rate of replication within a host, which usually increases the probability of its being transmitted to a new host, is balanced with its negative effect on the duration of the infectious period (May and Anderson 1983). If the pathogen is not virulent, it is unlikely to reach a high transmission rate. Conversely, a pathogen that replicates intensively in the host will have a higher transmission rate but over a shorter time because rapid host exploitation also means shorter host lifespan. Similarly to Achilles who, according to Homer, had to choose between a short and glorious life or a long but dull one, the pathogen has to evolve a strategy between causing long and mild infections or short and virulent infections (Alizon et al. 2009). If such a trade-off is at work, external factors can affect virulence evolution in a predictable way. For instance, the lower the base-line mortality of the host (independently of the disease), then the higher the pathogen virulence should be. This is so because the infectious period is reduced and the pathogen has to use up the host resources in a shorter time. There is also a growing interest in the host reaction to an infection, which can broadly be split into resistance (i.e. fighting the disease, which decreases both virulence and transmission) or tolerance phenomena (decreasing only the virulence, not the transmission rate) that affect parasite evolution (Raberg et al. 2009; Boots et al. 2009).
The density of susceptible hosts can also affect short-term evolutionary dynamics of virulence, as clearly shown by a recent evolutionary epidemiology framework that combines epidemiology and population genetics (Day and Proulx 2004). Indeed, early on during the course of an epidemic, most of the hosts are susceptible to infection so natural selection acts to favour strains with a high transmission rate (which happen to be more virulent according to the trade-off hypothesis). Once the disease has reached an endemic stage, the pool of susceptible hosts is smaller (hosts are either already infected, dead, or immunised) and natural selection then acts to favour strains that cause longer infections. In conclusion, virulence can thus be expected to vary over the course of an epidemic for rapidly evolving pathogens.
Another dimension of the model is that it does not concern itself with morbidity (at least not explicitly). Thus, symptoms like pain or injuries are not taken into account by the trade-off model and are implicitly integrated with other variables like host recovery and parasite transmission (Levin 1996). This assumption impacts on the ways in which virulence will be measured and operationalized. Whereas for doctors morbidity (illness) is a key feature of virulence, for evolutionary biologists or population biologists host’s pathological factors do not need to be taken into account when measuring virulence; what matters are effects that modify the pathogen’s fitness (i.e. that appear in the expression of R 0 ). In sum, the model rests on the idea that the pathogen transmission rate cannot increase beyond a certain point without at the same time inflicting damage to the host which would, in turn, be harmful to the pathogen by decreasing the duration of the infection. What matters for an evolutionary biologist is the fitness of an individual where the fitness of a parasite strain typically is given by the R 0 , i.e. the number of secondary infections. In other words, for a given parasite species, natural selection favours strains with the highest R 0 . This can explain why the highest possible level of virulence is not always the evolutionary stable (“optimal”) strategy to increase parasite’s fitness: increased transmission will indeed increase one component of parasite fitness R 0 (the transmission rate) but it will also decrease another component of R 0 that is the duration of the infection (through increased virulence) as the host is likely to die more rapidly from the infection. The balance between the two selective pressures (transmission favouring higher virulence and duration of infection favouring lower virulence) determines the evolutionary stable level of virulence.
Applying the Trade-Off Model to the 1918–19 Influenza Pandemic
Ewald’s early work on pathogen’s virulence and transmission developed a verbal theory for the trade-off model by comparing diseases with different transmission routes (Ewald 1983). His work was based on the concept of “cultural vectors” and on the assumption that parasites that do not rely on host mobility for transmission should evolve towards higher levels of virulence. In Ewald’s terminology, a cultural vector is “a set of characteristics that allow transmission from immobilized hosts to susceptible when at least one of the characteristics is some aspects of human cultures” (1994; 68; see also Ewald 1988). In the case of waterborne transmission, cultural vectors include contaminated bed sheets in hospitals, sewage systems carrying the pathogens, medical staff disposing of the contaminated water to water supplies, and so on. Waterborne diseases can become more virulent because they do not rely on host mobility for transmission (see Ewald 1994, 69), that is, the host can be isolated and still be highly contagious; a “healthy” host is not needed for transmission (in contrast with what was postulated by the conventional wisdom). Note that implicit in his reasoning is the idea that more virulent pathogens have a higher transmission because they produce more spores.
Applying the trade-off model to the case of the 1918 pandemic, Ewald argued that host proximity and population density were key elements in enhancing virulence. More precisely, he argued that the exceptionally high virulence resulted from rapid passages of the virus in soldiers, recruits and wounded people in hospitals during the war. Though a similar explanation had already been heralded in the 1930s–1940s, it had to be supplemented with an essential “evolutionary mechanism”: the classical explanation is based on the analogy with rapid passages of a viral strain through a series of animals (i.e. guinea pigs) in a laboratory that can enhance virulence (as Pasteur et al. (1994 [1881]) had experimentally demonstrated, see Mendelsohn (2002)). Ewald’s argument is that, just like biological vectors, cultural vectors enhance virulence by facilitating transmission. The central point about the serial passages is that it removes the “requirement that hosts be mobile to transmit their infections” (Ewald 1994, 115). Once this obstacle is lifted nothing (a priori) stands in the way of a steep increase in virulence. In a laboratory context, experimenters inoculate different animals with artificially selected viral strains; in the field, this selection process results from another cultural vector, namely the warfare conditions.
In the trenches, during the Great War, conditions were such that transmission was maximized and with it, the observed level of virulence. As postulated by the trade-off model, the density of the population (S) influences the level of observed virulence in a biological system (at least for short-term evolutionary dynamics). In this case, the high density resulted from the proximity of the soldiers in the trenches, in hospitals, on trains bringing soldiers to the front, and in military camps. In turn, this resulted in the unusual situation that immobilized individuals who normally should not be able to infect new people (because they would be isolated in a hospital) were now easily able to transmit the infections. Similarly, removing wounded soldiers from the trenches and bringing them to war hospitals facilitated transmission. The constant arrival of new susceptible individuals into the population through transport networks resulted in maintaining a high density of infected people; and as a consequence, an equally high level of virulence. Related to this is the idea that spatial structure in the host population affects virulence evolution. If hosts tend to have few contacts among them, e.g. because they live in isolation (the technical term to describe such a population is “viscous”), then a parasite has to keep its host alive sufficiently long enough to be transmitted. On the other hand, if the population is “well mixed”, host encounter rate is not an issue – as in the 1918–19 example – and parasites can afford to be more virulent (Boots and Sasaki 1999).
Some Problems with Ewald’s Account
Despite its theoretical appeal, some detected a number of problems in the explanation advanced by Ewald and with the trade-off model in general. For other evolutionary ecologists, Ewald’s cost-benefit argument is too adaptationist – i.e. virulence is depicted as being always adaptive for the parasite. As a consequence, “alternatives such as virulence being non-adaptive, or virulence being a consequence of short-sighted, within-host evolution of the parasite are ignored” (Bull and Levin 1994, 1470). Evolutionary theory states that virulence can be directly selected but it can also be coincidental with other infection or biological processes (Levin and Edén 1990), and in some cases it can be potentially maladaptive. This point connects to one of the usual critiques levered against the trade-off hypothesis, namely that it is very verbal and lacks empirical support (Levin and Bull 1994; Lipsitch and Moxon 1997). However, the lack of support largely comes from the difficulty of finding an appropriate biological system; arguably, when people have looked for a trade-off in a host parasite system that satisfies the assumptions of the theory they have found it (Alizon et al. 2009). There is actually a tendency to challenge the trade-off hypothesis using host-parasite systems that do not fit the underlying model (see Alizon and Michalakis (2011) for an illustration).
A second problem stems from the low level of transmissibility in influenza viruses and rate of pathogens’ reproduction. When taken into account, this concern weakens the claim that high transmission in the case of the 1918 pandemic has favoured high virulence because transmission was lower than with most infectious diseases. R 0 are typically variable but given Ewald’s argument it would be expected to find a high transmissibility rate between the virus and its hosts. In turn, this would support the claim that natural selection acted on transmission in ways that increased the overall level of virulence. Moreover, the trade-off assumes a homogeneous population and was developed for diseases transmitted by contact like influenza. However, a comparison of R 0 between the 1918–19 pandemic with other major disease outbreaks in recent history, or with influenza pandemics in general, does not reveal a significantly higher transmissibility in the case of the Spanish influenza. Calculations suggest that the basic reproductive rate of viruses during influenza pandemics ranges from 2 and 3 (Mills et al. 2004). In comparison, the reproductive rate during an outbreak of measles in England in 1947–1950 was between 13 and 14 secondary infections; a pertussis outbreak in Maryland (U.S.) in 1913 yielded a reproductive rate between 7 and 8; and a mumps outbreak in the Netherlands during the 1970s produced between 11 and 14 secondary infections in a wholly susceptible populations (Anderson and May 1991, 70). In the case of the 1918 pandemic, more recent calculations suggest that R0 was perhaps equal to 2 (Morse 2007, 7314). Finally, a recent article on the transmission of influenza in households during the pandemic (Fraser et al. 2011) used historical data and mathematical models to study the rate of transmission. The authors found a relatively low level of transmission between individuals and suggest that prior immunity to the virus should be considered. Though transmissibility may, theoretically, have been fostered so that the virus reached unprecedented virulence, the trade-off model alone does not fully explain why it was so deadly.
A third issue is the lack of empirical details in Ewald’s explanation of the steep increase in virulence circa 1918–19. To make his argument more compelling, Ewald needs additional data that accurately and empirically describe the environmental conditions in the trenches. For instance, how close were the troops? How many soldiers were there? And more importantly, what was the rate of transmission between hosts? If a similar study to Fraser et al. (2011) could be conducted on viral transmission in the trenches it would perhaps yield interesting insights into the changes of virulence. To date, no epidemiological data exists that could serve as a basis to model the dynamic patterns, however. Though Ewald’s account seems to suffer from a number of theoretical and empirical problems, it nevertheless supports the argument that properties other than those of the virus need to be taken into account and that without them we would are not able to fully understand the changes in virulence that occurred. As he remarked, progress towards the evolution of virulence “has largely been limited to improve understanding of the genetic mechanisms of antigenic changes and the influences of these changes and host immunity on the occurrence of epidemics” (Ewald 1991, 15). The recent work of microbiologist John Oxford on what we could call the “War Hypothesis” reinforces Ewald’s conclusion by feeding in some of the missing empirical and historical data.
The “War Hypothesis”
While many would agree that the Great War is a variable that must be included, in one way or another, in the broader explanation of the steep evolution of virulence of the 1918–19 pandemic, Ewald is convinced that the influenza pandemic was “caused evolutionarily by the war rather than being just coincidental with the war” (Ewald 1994, 115). The “War Hypothesis”, as we may call it, received new support from Oxford (2001; Reid et al. 2003; Oxford et al. 2005) who does not claim that the Great War caused the disease, evolutionarily or otherwise, but instead that the war created the right environment for the virus to become extremely deadly. When the 1918 pandemic broke out air travel was minimal and this suggests, according to Oxford, that “earlier ‘seeding’ has occurred” (2001, 1857). Taking an environmentally oriented approach to the evolution of virulence, Oxford and his colleagues argued that the 1918–19 pandemic originated in France in 1916 before going global 2 years later.Footnote 15 They did not postulate the evolutionary emergence of a mutant strain but rather that the ecological conditions facilitated the spread of a pre-existing influenza strain. Studying several epidemiological and medical reports of sporadic outbreaks of respiratory infections at the British base camp in the town of Etaples in Northern France in 1916, Oxford argued that the disease’s clinical picture maps very precisely onto the description of the 1918–19 influenza: not only were the 1916 respiratory diseases extremely deadly, but post-mortem examination revealed in most cases clear evidence of bronchopneumonia and histological analyses of lung tissues indicated “acute purulent bronchitis” (Oxford 2001, 1857).
In an article published a few years later (Oxford et al. 2005), Oxford and his colleagues took their examination of the situation one step further. Rejecting the possibility that “a particular virulence gene of influenza” could help to identify future pandemics, they argued that surveillance and detection of emerging influenza pandemics would be best served by understanding the contexts that give rise to pandemics, rather than by an analysis of genetic factors alone. In particular, concerning the 1918 pandemic, they noted that so far “there is no clear genetic indication of why this virus [the 1918 strain] was so virulent”. They also remarked that what is needed is a closer examination of the environmental and social conditions of the time such as population upheavals to explain the exceptional virulence. The authors asked specifically whether “the special circumstances engendered in the war itself have allowed or caused the emergence, evolution and spread of a pandemic virus” (Oxford et al. 2005, 941). For them, the “unprecedented circumstances” of the war in Europe were critical. Back in 1918, the Front was
a landscape that was contaminated with respiratory irritants such as chlorine and phosgene, and characterized by stress and overcrowding, the partial starvation of its civilians, and the opportunity for rapid “passages” of influenza in young soldiers would have provided the opportunity for small mutational charges throughout the viral genome […] could have been important factors in the evolution of the virus into a particularly virulent form (Oxford et al. 2002, 113).
The military camp of Etaples in France was subject to high traffic in 1916–1917. In addition to soldiers moving up to the Front and back, 230,000 sick and injured individuals were in the hospitals “at any given time”, making them overcrowded and allowing the virus plenty of opportunities for “rapid passages”. Overall, it is estimated that the region of Etaples hosted two million soldiers who camped there during the war, in addition to the six million others who occupied and fought in the trenches system that connected the English Channel with Switzerland (Oxford et al. 2005, 942). Secondly, as the camp had an “extensive piggery”, villagers could buy geese, ducks, and chickens, providing ideal conditions for the influenza virus to undergo antigenic shift. Thirdly, the extensive use of gases during the war (estimated at one hundred tons), some of which were mutagenic rendered the soldiers immunocompromised and more susceptible to influenza infections. Finally, demobilisation after the war sent the soldiers back home by boat or by train, and contributed to the spread of the disease by person-to-person contact all over the world (Oxford et al. 2005). Taken together, all these factors (overcrowding, being immunocompromised, pig-duck farming, demobilisation) created exceptional conditions for the virus to go pandemic. Ewald had noted that in the absence of a recreation of those circumstances it is unlikely that such a severe pandemic will happen again. What Oxford and his colleagues emphasized in their turn is that the appropriate response to a future pandemic cannot rest of putative virulence genes alone; one has also to consider the context that will allow the virus to spread in a pandemic fashion. At the same time that this ecological perspective was developed, another view on the sources of virulence was well underway in the United States.
Emerging Technologies
In the mid-twentieth century, leading British bacteriologist Wilson Smith, co-discoverer of the viral nature of influenza in humans in 1933,Footnote 16 doubted that the exceptional virulence could be linked to a particular genetic or molecular structure of the virus alone: “if we had the chance of getting a 1918–19 strain of the influenza virus now”, he said, “it is at least conceivable that, on comparing with the Asian strain, we might find no difference in intrinsic virulence at all, but the conditions in the human population during the two epidemic periods might have affected the degree of heterogeneity displayed by viruses possessed of the same intrinsic virulence” (1960, 77). His comments were intended to provide support to a paper delivered earlier by Edwin Kilbourne, an American virologist who specialised in influenza, who had argued that the greater virulence of the 1918 pandemic was due to a combination of “the emergence of a new antigenic type in a population with little specific immunity” and “the dislocation of and crowding of wartime which favoured not only dissemination and high dosage of virus but spread of bacterial pathogens to an unusual degree” (Kilbourne 1960, 74). Kilbourne argued that the “study of the host and his environment are more crucial to the interpretation of virulence than laboratory study of the virus itself” (1960, 71). Attempts to locate the cause of virulence inside specific genes or to relate them to other mobile, structural elements (i.e. plasmids) were met with scepticism by people like Wilson, Kilbourne, and Burnet who were interested in large-scale ecological processes and in the formation of evolutionary equilibriums between hosts and parasites. Ecologically minded biologists were also reacting against the growing place of molecular biology since the 1960s and its reductionist vision of the life process and the life sciences. This is how we can interpret Kilbourne who scornfully remarked that “ironically in this era of molecular biology, the control of no infectious disease has yet depended on understanding its molecular mechanisms” (1977, 1228).
In the early 1950s, scientific expeditions were organized to discover the remains of victims of the Spanish flu in the hope of finding traces of the virus. One of the expeditors was John Hultin (1925-), a pathologist from Sweden who immigrated to Iowa in 1949 to study medicine. As part of a project funded by the University of Iowa, he travelled in 1951 to a small Inuit village whose population was decimated by the 1918 pandemic, which had killed 70 people in a week, a loss amounting to 85 % of the inhabitants. Hoping to find preserved corpses buried in the permafrost hosting traces of the infectious organism, Hultin travelled to the Seward Peninsular of Alaska in a village known as Teller Mission (Taubenberger 2003). He extracted lung tissue from several bodies he exhumed from the village cemetery, but all attempts to culture remaining traces of the virus of influenza from these samples failed to give any result. Forty years later, in the context of the Human Genome Project, the idea of resurrecting the influenza virus surfaced again, this time powered by genomic technology.
Since the late 1990s, a renewed emphasis has been placed on the molecular, internal constituents of virulence. Newly developed technology and the availability of pathogenic viral and bacterial material have facilitated the development of this approach towards explaining infectiousness. This led, in 2005, to the publication of the complete influenza virus’ genomic map in both Nature and Science. Though all samples of the 1918 strain were thought to be long extinct and lost, bits of RNA of the virus were discovered and processed in order to generate the complete map of its genetic structure. After the discovery of frozen individuals killed by the 1918–19 pandemic and preserved in permafrost, scientists worked on the pathogenic mechanisms that possibly enabled the influenza virus to achieve unprecedented levels of virulence. Microbiologist Jeffrey Taubenberger of the National Institute of Allergies and Infectious Diseases in Washington led this work together with Terrence Tumpey from the Center for Disease Control in Atlanta. We now turn to this recent technological success and the difficulties of pinpointing any particular molecular feature of the flu virus of 1918 that could account for its exceptional virulence.
Traces of the Spanish Flu: From (Sero)archeology to PCR Amplification
In 1997 a U.S. lab-group based at the Armed Forces Institute of Pathology in Washington (D.C.), and led by molecular pathologist Jeffrey Taubenberger, published a piece in Science titled “Initial genetic characterization of the 1918–19 ʻSpanish’ influenza virus” (Taubenberger et al. 1997). The article provided a first and partial genetic map of the virus from “archival formalin-fixed, paraffin-embedded autopsy tissues of 1918 flu victims” (Taubenberger 2003, 42). The examined samples were kept at, and provided by, the National Tissue Repository of the Armed Forces Institute of Pathology. As several mutations in hemmagglutinin, especially on cleavage-sites, often contribute to the virulence (e.g. on influenza subtypes H5 and H7) by increasing the tissue tropism, it was hoped that the genetic make-up of the virus would provide insights into the virulence of the 1918 Spanish influenza pandemic. The goal of the project was “first, to discover where the 1918 influenza came from, and how it got into people, and second, whether there were any genetic features of the sequence that would give insight into the exceptional virulence of the strain” (Taubenberger 2003, 44).
This first publication of the team describes the technique used to obtain, amplify (PCR), and sequence the genetic material. The main finding of the paper, based on molecular phylogenetic analyses of gene segments, was that the 1918 pandemic was caused by a strain of H1N1 influenza virus, and that it was of avian origin (1997, 1795). In their first article, Taubenberger et al. randomly selected 28 cases of paraffin-embedded tissues collected from army servicemen who died during the pandemic for pathological review, searching for symptoms indicative of death by influenza. Most of the individuals examined died of secondary pulmonary infection, which was a common feature of victims of the pandemic. In effect, bacterial infection very often works together with the influenza virus in delineating the clinical picture of the disease. One case, indeed (1918 case1), could be linked to viral pneumonia and exhibited symptoms of acute pneumonia in the left lung combined with an acute form of bronchiolitis in the right lung, a pathological characteristic typical of a “primary viral pneumonia”.
Focusing on case1, researchers performed control amplification of reverse-transcribed genetics of the nine gene fragments of the 1918 virus using the technique of polymerase chain reaction (PCR). They then carried out phylogenetic analyses based on the gene sequences to reconstruct the genealogical relationships between these elements. It was concluded that the genetic sequence of this strain was different from every other influenza strain, and that it was more closely related to strains found in birds than in mammals (Taubenberger et al. 1997). This partial analysis of the genetic map of human influenza was soon followed by a complete sequencing of the hemmagglutinin gene (HA) – a gene long believed to be “pivotal” in the pathogenicity of influenza A viruses (Webster and Rott 1987; see also Cox and Bender 1995). This gene codes for a protein located on the surface of the virus that plays a crucial role in allowing the virus to bind to host cells. If the virus is able to spread to another species this means it has somehow (through antigenic drift) acquired a new protein that enables it to bind on a different receptor. However, the team did not identify a mutation of the cleave site of the hemmagglutinin gene (Reid et al. 2001; Taubenberger et al. 1997).
Two years later, the team published another article on the “Origin and evolution of the 1918 ‘Spanish’ influenza virus hemmagglutinin gene” (Reid, Fanning, Hultin, and Taubenberger 1999). Johan Hultin, the pathologist who attempted to find traces of the influenza virus in Alaska in the early 1950s, was among the authors of the study. After reading the 1997 Science paper, Hultin wrote a letter to Taubenberger offering to return to Brevig Mission to look for samples of people who had died of the flu (Taubenberger 2003, 43). Against all odds, Hultin was successful. After he received the approval of Taubenberger he set out to Alaska for a second time and in August 1997 he found in situ frozen lung biopsies. Once in the village, he was granted permission from the council to dig the graveyard again; with the help of a few villagers and after 4 days work, he unearthed the body of a 30 year-old woman whom he called “Lucy”. Opening up her chest he found two frozen lungs that he immediately sent to Taubenberger’s laboratory in Washington, along with some tissues taken from three other frozen corpses (Berche 2012).
Reid et al. (1999) reported on the full sequence of the hemmagglutinin gene using RNA fragments from case1 discussed in the first article. They investigated three case histories to find evidence of influenza RNA. The first one was a 21 year-old man who died at Fort Jackson in South Carolina. Pathological records indicate he had pneumonia and influenza symptoms; he was admitted to the camp hospital on September 20th 1918 and died within 6 days. The autopsy records also show that his left lung suffered from an acute and fatal attack of pneumonia, whereas his right one showed acute bronchiolitis and alveolitis – a clear sign of influenza infection. No RNA was found in the left lung. However, the team performed a minute microscopic analysis on the paraffin-embedded tissue of the right lung and tissues tested positive for influenza RNA. The fragments of five genes were sequenced, amplified through PCR technique and then determined. The second case was also a male soldier, this one 30 years old and based at Camp Upton in the State of New York. He was admitted to hospital with pneumonia and died within 3 days on the 23rd of September 1918. Microscopic examination of his lungs by Taubenberger and his team revealed acute pulmonary oedema and acute bronchopneumonia. Formalin-fixed, paraffin-embedded samples of lung tissues tested positive for influenza RNA, the sequence of which was no longer than 150 nucleotides. The third case history was the one found by Hultin in Brevig Mission, Alaska.
Using the sequences of these three case histories, the Washington-based team worked out the genealogical relationships between them. Their analysis reasserted that the virus that caused the pandemic was avian in nature and that it entered human populations between 1900 and 1915, following the modification of the binding site on the HA protein. In 2005, Taubenberger and Tumpey published two separate articles in Nature and Science: the first provided the complete genomic sequence of the 1918 influenza virus and the second revealed the methods used to artificially reconstruct it. Yet, even before the complete genomic map of the virus was made available, it became unclear whether the genes of the influenza virus had indeed disclosed the causes of its exceptional virulence (see Taubenberger et al. 2001). Moreover, their argument of a likely avian origin of the virus was criticized.
A Missing Mutation and the Limits of Genomic Analyses
Efforts to sequence the virus that caused the 1918–19 influenza pandemic were motivated by the possibility of understanding the genetic origin and virulence of such an organism. While this work allowed for a more precise characterization of the hemagglutinin, neuraminidase, matrix, and nucleoprotein gene segments from a functional point of view, it is less clear, however, whether the first goal was achieved. In effect, the Washington team reported that a cleavage-site mutation on the hemmagglutinin gene that played a crucial role in the virulence of the Hong Kong pandemic in 1968 was not found in the strain obtained from the South Carolina case. Sequencing the specific cleavage site in the RNA of the virus obtained from the Brevig Mission case and New York case also confirmed that this mutation was absent. Inquiring into this mutation site (hemmagglutinin) – understood as a key determinant of virulence – was a central motivation of Taubenberger’s work as it would have “offered an appealing explanation of the 1918’s flu virulence” (Taubenberger 2003, 45). Yet Taubenberger was forced to recognize that “the 1918 strain (as confirmed by all three cases) does not possess a mutation at this site” (Ibid.; see also Reid et al. 1999; Stevens et al. 2004). In the light of this conclusion, virologist and influenza expert Robert Webster wrote that the secret of the Spanish influenza will “remain elusive”. Webster commented that such “biological properties” [i.e. virulence] may “not be resolved” and suggested that the results of the sequencing project could only provide a partial explanation of this phenomenon. Indeed, for him “the entire gene sequence is unlikely to reveal the secret of the high pathogenicity of the 1918 Spanish virus” (Webster 1999, 1165). While Taubenberger’s paper ends with some remarks about the complex, likely polygenic, nature of virulence determinants in a particular strain, it also concludes – contra Webster – with the hope that more sequencing would “shed additional light on the nature of the 1918 influenza virus” (Reid et al. 1999, 1656).
Another molecular explanation of the 1918–19 flu pandemic emerged in 1998 from another research team. Virologists Hideo Goto and Yoshihiro Kawaoka published a paper in the Proceedings of the National Academy of Science on a novel mechanism for the acquisition of virulence by human influenza A viruses. There, they argued that a change in another major protein – neuraminidase – able to increase the cleavage of HA could bring about higher levels of virulence. In fact, Goto and Kawaoka even suggested that a change in a single amino-acid sequestering plasminogen might facilitate the cleavage of NA. The authors were cautious, however, stating they “do not conclude that single mutation will convert nonplasminogen-binding NAs to efficient plasminogen binders, thus rendering the virus highly virulent” (1998, 10228). Yet, they acknowledged at the same time that it is “tempting to speculate that the 1918 pandemic strain […] may have acquired its unprecedented virulence from the mechanism we describe” (Ibid). But such a change in amino-acid was also absent (or at least not observed) in the 1918 neuraminidase sequence (Taubenberger 2003, 45; 1988; see also Reid et al. 2000; Kawaoka and Watanabe 2011). Also, similar to Taubenberger, Goto and Kawaoka concluded with a plea for “further sequencing”, in order “to address the issue of its [the 1918–19 pandemic] unprecedented virulence” (Goto and Kawaoka 1998, 10228).
In 2004, both Taubenberger and Tumpey acknowledged the lack of evidence provided by the molecular structure of the virus to explain its virulence:
Sequence analysis of the 1918 influenza virus from fixed and frozen lung tissue has provided molecular characterization and phylogenetic analysis of this strain. The complete coding sequence of the 1918 nonstructural (NS), hemagglutinin (HA), neuraminidase (NA), and matrix (M) genes have been determined; however, the sequences of these genes did not reveal features that could account for its high virulence” (Tumpey et al. 2004; emphasis added).
And yet, despite evidence for an absence, there seems to be something particular about the structure of the HA protein that contributes to an enhanced level of virulence (Morange 2005). Indeed, using a mouse model, another team of molecular pathologists (Kobasa et al. 2004) showed that when the HA protein taken from the 1918 viral strain is inserted into mice it confers high pathogenicity and facilitates lung infections. For instance, infected mice show 39,000 times more virus particles after infection with the 1918 strain than with other viral strains like the Texas virus, and infected mice died after 6 days following infection with the 1918 strain, while all survived when infected with the Texas virus (von Bubnoff 2005, 794). The particular structure of the protein responsible for such pathological effect remains to be found, however, and it is unclear whether similar effects could hold true in humans as well.
Evolutionary Explanations in Emerging Diseases and Changes in Virulence
As we have described, Taubenberger’s team provided the first molecular characterisation of the Spanish influenza organism based on the construction of phylogenetic trees of 9 of the 11 RNA-polymerase genes of the 1918–19 virus (Taubenberger 2005). The authors of this research project that spanned several years concluded that the virus did not originate from gene reshuffling (or reassortment) but rather that it jumped from birds to humans shortly before the onset of the pandemic. The virus was thus of avian origin. However, their interpretation of the similarity by descent, and thus of the genealogical relationships between the 1918 virus and today’s avian viruses was disputed (Gibbs and Gibbs 2006; Antonovics et al. 2006).
As the current head of the Viral Pathogenesis and Evolution Section at the National Institute for Allergy and Infectious Diseases, Taubenberger’s work is underpinned by evolutionary considerations. But what aspects of his work exactly are evolutionary or Darwinian? Philosopher Michael Ruse has long pointed out that the term “Darwinism” carries two broad meanings. It can be used firstly in a metaphysical sense to characterize change, development and transformation in the natural world. In this sense, the concept of Darwinism is older than Darwin himself. Another sense of Darwinism is important to acknowledge. In this second sense, Darwinism is a scientific notion that emerges in the work of naturalist Charles Darwin and refers to the fact of evolution, the paths (phylogenies) of evolution, and the mechanism (natural selection) of evolution (Ruse 1992, 77). This distinction between path and mechanism maps on the more traditional distinction between patterns and processes in evolutionary biology mentioned above. The research of Taubenberger and Ewald – and more generally molecular pathology and evolutionary ecology – displays these two aspects of Darwinian theory. Arguably, both accept evolution as a “fact”. However, the former is more interested in the “patterns” of evolution and uses evolutionary thinking to unravel the biological (including genetic) and adaptive processes that led to an increase in virulence. In contrast, Ewald focuses on the “process” of evolution – natural selection – as it occurred in various environments and populations of hosts and pathogens. As described above, Taubenberger’s research focuses on precise and minute description of the small steps that allow viruses to infect more than one species; this work painstakingly tracks changes in nucleotides and charts the genealogical relationships between several strains of influenza. Ewald and Oxford, in contrast, take a broader view and ask why those mutations were selected, what were the selective pressures that drove them to be passed on and conserved in the gene pool, and especially, what is the role of the milieu, largely understood, in shaping virulence.
Though the centrality of the concept of natural selection is not really in dispute here, the ways in which Taubenberger and Ewald (and other evolutionary ecologists) understand these processes differs significantly on one important point: whereas the former describes the small incremental steps leading to the high, observable level of virulence, the latter looks for a plausible, eventually testable evolutionary scenario leading to the accumulation and conservation of these small, gradual changes. In other words, the second approach, the ecological one, seeks not only to describe organic changes leading to the formation of new viral strains, for example, but also attempts to give an account of the adaptive value of these transformations in the particular milieu in which the microorganisms lived, reproduced and eventually died. These two components of evolutionary theory – patterns and processes – are well known in the history of biology. Evolutionary ecologists nowadays might want to argue that Taubenberger is primarily interested in constructing and comparing distinct phylogenetic trees, no matter what the significance of their (evolutionary) relationship may be.Footnote 17 We think that the two aspects of evolutionary theory discussed here, however, reflect more broadly the existence of two distinct styles of scientific practices in biomedicine. The difficulty in addressing both aspects of the theory at the same time is indicative of a genuine tension between distinct explanatory strategies where knowledge claims are made according to different assumptions as to what counts as explanatory.
A Note on the Plurality of the Scientific Styles of Practice
Confronted with the lack of evidence supporting a molecular explanation, and in the light of the limitations of the environmental-ecological account, one could have expected researchers to seek support in each other’s work in order to complement their researches, and to move beyond the limitations of their own methodologies and research paradigms. Yet it is striking to note that Reid, Taubenberger et al. (1999), on the one hand, and Goto and Kawaoka (1998), on the other, reached a conclusion diametrically opposed to that of Webster and also Ewald: for the former, in order to explain better the influenza pandemic, more genomic sequencing is needed. Instead of considering other possible explanations of the exceptional virulence (i.e. ecological explanations) they persist in their attempt to provide a complete and satisfying explanation within a single explanatory framework.
At this point, a few remarks are in order. Firstly, and from a broad sociological point of view, this may just be a sign of our times: sequencing genetic material is an effective, and now rather inexpensive, way of obtaining prestigious research grants. Proposals in genomics, synthetic biology, and other cognate fields with a strong engineering approach to biology can highlight potential findings and even future applications, some of which are likely to be patentable and thus rapidly rentable from a financial point of view. In brief, promoting more sequencing is likely to provide additional research money. While this may be a reason why Taubenberger’s team value more genetic sequencing other reasons of a more epistemological and historical nature must also be envisaged.
A second reason to consider has to do with what historians and philosophers of science have called a scientific “style of practice” (Keating and Cambrosio 2007). Derivative of Ian Hacking’s concept of “styles of reasoning” – itself inspired by Alistair Crombie’s “style of scientific thinking in the European tradition”– (Hacking 1992; Crombie 1994), the notion of “style” typically refers to the historical formation of distinctive practices and methodologies in science. Styles frame what counts as evidence, relevant questions to ask, truth-value, and sound explanation in distinct research and/or cultural contexts. Alongside the development of individual styles of practice one finds the emergence of new standards for measurements, objectivity, proof, and so on (Hacking 1992). Though styles are flexible they are not loose or relativist categories; they admit rules, systems of norms, stabilization techniques, and methods of justification. As they progressively become stabilised over time and entrenched within scientific activities, however, the very existence of styles of reasoning and their historical development become taken for granted. While the notion of style is often employed to analyse scientific controversies (Amsterdamska 2004; Fujimura and Chou 1994), it is interesting to note here that the two styles at play in the present case study have grown in relative ignorance of each other. Going back to the missing mutations, we can see that even though Taubenberger’s programme did not provide the answers it sought it could not be halted hastily, especially after gathering immense publicity and funding. On the contrary, it is expected that these scientists, working within their style of practice, continue to do so until all possibilities of finding the key to the exceptional virulence have been looked at and examined in detail. From this point of view, their persistence in seeking a complete molecular explanation makes sense – even if, from a public health and biosecurity point of view, their research raises ethical concerns about the development of dual-use technologies (Rappert 2007). Moreover, the results obtained on the biology of influenza A viruses and the methods developed by Taubenberger and his team now enable worldwide researchers to better understand the molecular differences between various influenza strains.
What appears as a sign of determination in pursuing a research objective can also reflect a lack of communication between distinct scientific communities, the problems of interdisciplinary work, the self-containment of styles of scientific practice, and/or the resistance offered by epistemological obstacles. The current gap between ecological and molecular explanations, as it emerged in the present case, may be due to the fact that functional explanations such as those constructed in molecular biology tend to appear “self-sufficient”, as historian and biologist Michel Morange recently put it (2011). This sort of epistemological obstacle means that, for many, there is no (obvious) need to complement molecular explanations with ecological considerations. To say that integration between mathematical modelling and molecular microbiological approaches has failed in this case would be going too far, however. Indeed, integration of ecological and molecular approaches of virulence evolution has not even been seriously attempted so far. Also, it would be misleading to suggest that molecular pathologists wholly ignore the environmental perspectives on virulence evolution and emerging diseases.Footnote 18 Yet when they do take them into account, the result does not necessarily amount to a better integration of data, theories, or methods but reveals, instead, the heights of disciplinary boundaries and the valuing of one style of practice over another. For example, in one of the last publications of Taubenberger and his colleagues at the National Institute of Health, the authors concluded that the diminution of severity of influenza pandemics over time “is surely due in part to advances in medicine and public health, but it may also reflect viral evolutionary choices that favor optimal transmissibility with minimal pathogenicity - a virus that kills its host too fast or sends them to bed is not optimally transmissible” (Morens et al. 2009, 229; emphasis added). In other words, the biological interests of the virus will best be served by evolving lower virulence over time in order to facilitate transmission to new hosts, an explanation that rests on the conventional wisdom rejected by most evolutionary ecologists who advocate the theoretical trade-off model but that is still defended by some microbiologists. This may come as a surprise given that Anthony Fauci, on of the authors of the paper, and current head of the National Institute for Allergy and Infectious Diseases, has long criticized this view (see Fauci 2005). It shows, however, that branches of sciences in which the same problem is addressed, through distinct methodologies, can be surprisingly disconnected and separated by epistemic gaps, professional or institutional barriers. In other words, integration is no easy goal to achieve.
Problems within the molecular style of practice, however, are not only epistemological but also ethical and social. The publication of the whole sequence of the 1918–19 strain in 2005 sparked lively debates among scientists and the public as it raised concerns as to whether it was safe to publish the methodology used to resurrect the pathogen (Rappert 2007; Selgelid 2005). What if someone with nefarious intentions reconstructs the virus? How likely is it that this genetic information be used for harmful purposes? What if, by accident or not, the virus escapes into the environment? For some, like biologist Richard H. Ebright from Rutgers University, “there is a risk, verging on inevitability, of accidental release of the virus” but “there is also a risk of deliberate release of the virus”.Footnote 19 Yet others argued that the work of Taubenberger and Tumpey was entirely legitimate and could be applied to other areas and problems in virology such as the H5N1 pandemic and “could have an immediate impact by helping scientists focus on detecting changes in the evolving H5N1 virus that might make widespread transmission among humans more likely”.Footnote 20
The case of the Spanish influenza pandemic is today a classical example of a technology that has the potential for “dual-use” research (i.e. it could help to understand the disease and fight it, but it also could be used to disseminate it further in a population). A recent case of potential dual-use consequences in influenza research involving a group of researchers led by Ron Fouchier in the Netherlands and by Yoshihiro Kawaoka, from Madison, in the U.S, led in January 2012 to a 60 day suspension of research on influenza and virulent diseases, following consensus to delay publication.Footnote 21 Both teams had submitted a paper, to Science and Nature respectively, describing the methodology employed to artificially render an H5N1 influenza strain transmissible between ferrets (which is, arguably, a reliable indicator of possible transmission to humans) due to a mutation on the hemmagglutinin protein. Both studies have now been published (Imai et al. 2012; Herfst et al. 2012). As an aside, it is interesting to see that these two studies, although undoubtedly driven by molecular biology questions, were based on classical approaches in evolutionary ecology known as serial passage experiments (Ebert 1998).
In the early 1990s, the Institute of Medicine’s report on Microbial Threats (Lederberg, Shope, and Oaks 1992) and Stephen Morse’s Emerging Viruses (Morse 1993) have emphasized how emerging infectious diseases are posing a renewed threat to public health that needs to be addressed on a global scale, from the combined perspective of ecological and molecular approaches. The concept of emerging diseases has helped focus international efforts to contain infectious diseases within well-defined geographical and temporal limitations. With the (re)creation of the 1918–19 influenza strain and others (e.g. Yersinia pestis, polio virus, H5N1), a different form of biological threat arises and it requires different political, institutional, and legal response mechanisms. Indeed, while the threat of emerging infections was mostly perceived as coming from outside Northern-hemisphere countries, it now appears to be growing from within the heartland of Western countries itself. Instead of stressing possible disease invasions in previously unexposed countries (or with only low incidence of a particular disease), recently developed technologies in synthetic biology and genomics have opened-up the possibility to artificially create new diseases, or to resurrect old ones such as the plague, the influenza strain responsible for the 1918 pandemic, and the polio virus (Bos et al. 2011; Taubenberger 2005; Tumpey et al. 2005; Celo et al. 2002; Rosengard et al. 2002). On the other hand, it might be argued that new variants appear constantly and the risk of a laboratory accident might be comparable to what happens naturally in the field. Moreover, while dual-use is a characteristic of most life sciences nowadays (Atlas 2009), only a small number of experiments and experimental practices are, overall, seen as posing real threats to public health and global security (for a recent analysis see Aucouturier 2011; Morens et al. 2012). Finally, it is worth noting that dual-use technologies – like scientific research more generally – are often characterized by unexpected findings such as, for instance, the accidental discovery that a modified virus injected into mice was lethal to otherwise vaccinated animals (Jackson et al. 2001). As is often the case in science, the experimental system designed to answer certain questions opens-up theoretical and practical possibilities that could sometimes not be envisaged at the outset (Rheinberger 1997). If unpredictability and unforeseen results are truly the essence of scientific research, dual-use technologies are then an unavoidable trade-off to deal with, a point that reinforces the need to develop appropriate governance responses to biomedical research programmes on pathogens and potentially pathogenic organisms (Méthot 2014). More generally, those new research avenues underline the need for the development of a “culture of responsibility” (NSABB 2011) in the life sciences, that is, a new ethos to address and balance questions of biosecurity and risk with scientific autonomy and progress, among others.
Concluding Remarks
The two most important glycoproteins allowing influenza viruses to invade host tissues – hemmagglutinin (HA) and neuraminidase (NA) – were significant molecular determinants of the virulence of influenza pandemics in 1957 and 1968, and can yield potentially pathogenic effects when inserted into some animal models. Considerable explanatory power was placed on these special proteins that seemed to provide a first-hand, adequate, simple and certainly elegant mechanism to account for the exceptional virulence of the 1918 pandemic. Indeed, the “most popular theory” was that the 1918 virus had “unique pathogenic properties, most likely encoded within the hemagglutinin protein” (Holmes 2004). Identifying a molecular and genetic basis of virulence could not only provide a window into the most devastating epidemic of modern times, but could also help to prevent and predict those to come. Overall, the remarkable technological success – i.e. the retrieving and sequencing of the 1918 avian virus – promised nothing less than to unlock one of the oldest and well-kept secrets in the whole of medical history. However, after sequencing the genome of the 1918 viral strain that killed perhaps up to 40 million people according to the WHO estimates, both factors were found to be lacking in the killer strain.
One might wonder about the extent to which it is possible to generalize from the example of the Spanish influenza pandemic to other cases. Researchers on ancient pathogens using high throughput technologies have recently claimed to identify the causal organism of the Black Death (Yersinia pestis) in the fourteenth century and the sources of its virulence in the form of a single plasmid (Schuenemann et al. 2001). However, determining why an organism is pathogenic or what makes it a pathogen is not straightforward and is rarely based on a specific structural characteristic alone (Méthot 2012c). As microbiologist Charles Nicolle once said (1930), virulence is the expression of a “mosaic of powers” resulting from a constellation of factors that are irreducible to any particular structure and must be understood against a broad biological and even historical background. It is interesting to note, therefore, that the same team went on to revise its position in a subsequent article by pointing out the inherent limitations of molecular-oriented explanation and, furthermore, emphasized the need to widen the explanation and integrate ecological factors as well. They write:
Regardless, although no extant Y. pestis strain possesses the same genetic profile as our ancient organism, our data suggest that few changes in known virulence-associated genes have accrued in the organism’s 660 years of evolution as a human pathogen, further suggesting that its perceived increased virulence in history may not be due to novel fixed point mutations detectable via the analytical approach described here. At our current resolution, we posit that molecular changes in pathogens are but one component of a constellation of factors contributing to changing infectious disease prevalence and severity, where genetics of the host population, climate, vector dynamics, social conditions and synergistic interactions with concurrent diseases should be foremost in discussions of population susceptibility to infectious disease and host–pathogen relationships with reference to Y. pestis infections (Bos et al. 2011; emphasis added).
In sum, the study of Bos et al. (2011) did not reveal any significant genetic or evolutionary change in 600 years that could explain the virulence of plague in the fourteenth century. As a consequence, they argue that a molecular approach only provides an incomplete picture when applied in isolation, and that a complementary ecological perspective is needed. More precisely, the more recent study emphasizes that a full understanding of the evolution of virulence requires a multi-dimensional framework that encompasses host resistance, ecological factors, and the interactions between the different diseases occuring in a well-defined geographical area over a specific time period. To go beyond the limitations of analytical approaches that investigate one disease at a time, a synthetic and global approach is necessary in order to understand more broadly the evolution of emerging diseases that compose the past, present, and future of any “pathocenosis” (Grmek 1969).
To conclude, our analyses of the case of the Spanish influenza pandemic show that there is an irreducible tension in studying the phenomenon of virulence comparatively across the sciences: one can either look for determinants of pathogenic power analytically, that is from within microorganisms, or synthetically at the level of host-pathogen interactions in a given ecological environment. Both approaches face limitations and neither appears to be sufficient to account for a complex phenomenon like the 1918–19 influenza pandemic. Besides, each level of analysis has its own ways of characterizing the nature and causes of virulence (and pathogenicity), and both lead to the development of different measures to prevent or to treat emerging infectious diseases. While ecological approaches contribute to the establishment of national and international programmes intended to increase detection and surveillance in emerging infections on a global scale, monitoring changes in virulence to prevent pandemics worldwide, molecular approaches facilitate the development of biomedical tools, such as vaccines or antibiotics, to fight infectious diseases either by reducing their pathogenic power, and/or by enhancing individual and group (or herd) immunity. Yet both perspectives can also work together: molecular phylogenies can provide evidence regarding the likely origins of future pandemics and help to channel the attention of a detection network onto particularly sensitive sites (see Morens et al. 2012). A better integration of ecological and molecular approaches would thus benefit public health medicine by providing stronger theoretical approaches and empirical mechanistic models to understand, manage and perhaps even predict future influenza pandemics.
Notes
- 1.
Here we use the term ‘parasite’ in its broad (ecological) sense, which encompasses both micro-parasites (viruses, bacteria, protozoa) and macro-parasites (e.g. worms).
- 2.
On the concept of pathogen, see Méthot, P.O. and Alizon, S. (forthcoming).
- 3.
- 4.
- 5.
See the recent special issue on integration in Studies in History and Philosophy of the Biological and Biomedical Sciences (2013).
- 6.
- 7.
Patterns derive from processes. The former can be described as the “study of order in nature” while the second refers to “mechanisms generating and maintaining this order” (Chapleau, Johansen, and Williamson 1988, 136).
- 8.
Note that, for historical reasons, in the phytopathology literature the virulence used to refer to the ability of the pathogen to infect a plant (i.e. a qualitative trait). Since 2001 the American Phytopathological Society has decided to use the term virulence to refer to the damage done to the host and the term pathogenicity for the ability to infect the plant but few researchers have adopted it. In a way, the debate between two fields (evolutionary ecology and molecular biology) has already happened within one of the fields (see Shapiro-Ilan et al. 2005; Thomas and Elkinton 2004; Shaner et al. 1992).
- 9.
On the origins of the trade-off model in May’s work, see Méthot (2012a).
- 10.
Before the formulation of the trade-off model, Macfalane Burnet declared: “there are no virulence genes as such” (1960, 1).
- 11.
Survival and reproduction often interact in a non-linear way. For a discussion on the epistemological aspects of the concept of fitness, see Bouchard (2004).
- 12.
On the history of the 1918 influenza pandemic, see Barry (2004a), Johnson and Mueller (2002), van Helvoort (1993), Crosby (1989), and Burnet and Clark (1942). For a short but informative “chronicle” of influenza pandemics, see Beveridge (1992), and for a detailed scientific account of the biology of influenza see Stuart-Harris (1953).
For recent histories on flu, see Bresalier, M. 2013 and Taubenberger and Morens (2010).
For a recent account of influenza pandemics, see Honigsbaum, M. A. 2013.
- 13.
The reference to Spain is due to the fact the publication of medical reports on influenza was authorized in Spain during the war, in contrast to other countries at war. As a consequence, the disease became associated with Spain that was subsequently blamed for it and considered responsible. One of the first papers to appear in London Times (June 1918) was titled “The Spanish Influenza – a sufferer’s symptoms” (in Johnson 2006, 37). Like syphilis, the Spanish influenza received other names in other countries, however. For instance, it was called the “Swiss flu” in France.
- 14.
For instance, Langford (2005) argues that the flu pandemic came from China.
- 15.
This research was later criticized in turn by Barry (2004b) who argued that the influenza pandemic originated in Kansas and was taken to Europe by U.S. soldiers in 1918.
- 16.
Until the late 1920s and early 1930s, and the work of Olitsky and Gates and Richard Shope, most bacteriologists believed that the causative organism of influenza was the Gram negative Pfeiffer bacillus (Haemophilius influenza) isolated by German scientist Richard Pfeiffer in 1892. However, the causal role of this organism in influenza aetiology was also disputed, particularly during the 1918–19 pandemic (Witte 2003). The discover of the viral nature of influenza was made by Smith, Andrewes, and Laidlaw in 1933 (Smith et al. 1933).
- 17.
- 18.
For instance, John Oxford collaborated on a paper with Reid, Taubenberger and several others in 2003 (Reid, Janczewski, Lourens et al. 2003).
- 19.
From New York Times article by Gina Kolate (2005) which can be accessed at http://www.nytimes.com/2005/10/06/health/06flu.html?pagewanted=all
- 20.
This statement was jointly issued by Fauci and Gerberding. See Kolate (2005).
- 21.
References
Alizon S (2014) Evolution of disease virulence. In: Losos J (ed) Oxford bibliographies in evolutionary biology. Oxford University Press, New York, 01/13/2014, doi:10.1093/OBO/9780199941728-0018
Alizon S, Michalakis Y (2011) The transmission-virulence trade-off and superinfection: a comment to Smith. Evolution 65:3633–3638
Alizon S, van Baalen M (2005) Emergence of a convex trade-off between transmission and virulence. Am Nat 165:E155–E167
Alizon S, Hurford A, Mideo N, Van Baalen M (2009) Virulence model and the trade-off hypothesis: history, current state of affairs and the future. J Evol Biol 22:245–259
Amsterdamska O (2004) Achieving disbelief: thought styles, microbial variation, and American and British epidemiology, 1900–1940. Stud Hist Philos Biol Biomed Sci 35(3):483–507
Anderson RM, May RM (1982) Coevolution of hosts and parasites. Parasitology 85:411–426
Anderson RM, May RM (1991) Infectious diseases of humans: dynamics and control. Oxford University Press, Oxford
Antia R, Regoes RR, Koella JC, Bergstrom CT (2003) The role of evolution in the emergence of infectious diseases. Nature 426:658–661
Antonovics J, Hood ME, Baker CH (2006) Was the 1918 flu avian in origin? Nature 440:E9
Atlas R (2009) Responsible conduct by life scientists in an age of terrorism. Sci Eng Ethics 15(3):293–301
Aucouturier E (2011) La guerre biologique: perspective historique et critique. PhD thesis, Université Paris1 (Panthéon-Sorbonne)
Ball GH (1943) Parasitism and evolution. Am Nat 77(771):345–367
Barry JM (2004a) The great influenza: the epic story of the deadliest plague in history. New York, Viking Books
Barry JM (2004b) The site of the origin of the 1918 influenza pandemic and its public health implications. J Transl Med 2(3)
Berche P (2012) Faut-il encore avoir peur de la grippe? Histoire des pandémies. Odile Jacob, Paris
Bergstrom C (2008) An evolutionary biologist considers the virulence of emerging infectious diseases. Nature Journal Club. 19th May, p 261
Beveridge WIB (1992) The chronicle of influenza pandemics. Hist Philos Life Sci 13:223–235
Boots M, Best A, Miller MR, White A (2009) The role of ecological feedbacks in the evolution of host defence: what does theory tell us? Philos Trans R Soc Lond B Biol Sci 364:27–36
Boots M, Sasaki A (1999) ‘Small worlds’ and the evolution of virulence: infection occurs locally not at a distance. Proc R Soc Lond B 266:1933–1938
Bos KB, Schuenemann VJ, Golding GB, Burbano HA et al (2011) A draft genome of Yersinia pestis from victims of the Black Death. Nature 478:506–510
Bouchard F (2004) Evolution, fitness, and the struggle for persistence. PhD. thesis, Duke University
Bresalier M (2013) A short history of flu. Bloomsbury Academic, London
Bull JJ (1994) Virulence. Evolution 48(5):1423–1437
Bull JJ, Levin BR (1994) Parasites on the move. Science 265:1469–1470
Burnet FM (1946) Virus as organism. Evolutionary and ecological aspects of some human virus diseases. Harvard University Press, Cambridge, MA
Burnet FM (1953) The future of medical research. The Lancet i:103–108
Burnet FM (1960) Chairman’s opening remarks. In: Wolstenholme GE, O’Connor CM (eds) Virus, virulence and pathogenicity. Churchill, London, pp 1–2
Burnet FM, Clark E (1942) Influenza, Monographs from the Walter and Eliza Hall Institute of Research in Pathology and Medicine. Macmillan Company, Melbourne
Bush RM (2007) Influenza evolution. In: Tybayrenc M (ed) Encyclopedic guide to infectious disease research: modern methodologies. Wiley, Hoboken, pp 199–214
Castillo-Salgado C (2010) Trends and directions of global public health surveillance. Epidemiol Rev 32:93–109
Celo J, Paul AV, Wimmer E (2002) Chemical synthesis of poliovirus cDNA: generation of infectious virus in the absence of a natural template. Science express. Report. www.sciencexpress.org
Chapleau F, Johansen PH, Williamson M (1988) The distinction between pattern and process in evolutionary biology: the use and abuse of the term ‘strategy’. Oikos 53(1):136–138
Cockburn TA (1963) The evolution and eradication of infectious diseases. Johns Hopkins Press, Baltimore
Cox N, Bender CA (1995) The molecular epidemiology of influenza viruses. Virology 6:359–370
Crombie AC (1994) Styles of scientific thinking in the European tradition: the history of argumentation and explanation especially in the mathematical and biomedical sciences and arts. Duckworth, London
Crosby AW (1989) America’s forgotten pandemic. The Influenza of 1918. Cambridge University Press, Cambridge
Day T, Proulx SR (2004) A general theory for evolutionary dynamics of virulence. Am Nat 163(4):E41–E63
Diekmann O, Heesterbeek JAP (2000) Mathematical epidemiology of infectious diseases. Model building, analysis, and interpretation. Wiley, New York
Ebert D (1998) Experimental evolution of parasites. Science 282:1432–1435. doi:10.1126/science.282.5393.1432
Ewald PW (1983) Host-parasite relations, vectors and the evolution of disease severity. Annu Rev Ecol Syst 14:465–485
Ewald PW (1988) Cultural vectors, virulence, and the emergence of evolutionary epidemiology. Oxf Surv Evol Biol 5:215–245
Ewald PW (1991) Transmission modes and the evolution of virulence, with special reference to cholera, influenza, and AIDS. Hum Nat 2(1):1–11
Ewald PW (1994) Evolution of infectious disease. Oxford University Press, Oxford
Ewald PW (1996) Guarding against the most dangerous emerging pathogens: insights from evolutionary biology. Emerg Infect Dis 2:245–257
Fantini B (1993) Les organisations sanitaires internationales face à l’émergence des maladies infectieuses nouvelles. Hist Philos Life Sci 15:435–457
Farmer P (1996) Social inequalities and emerging diseases. Emerg Infect Dis 2(4):259–269
Fauci AS (2000) Infectious diseases: consideration for 21st century. Clin Infect Dis 32:675–685
Fauci AS (2005) Emerging and re-emerging infectious diseases: the perpetual challenge, Robert H. Ebert memorial lecture. Milbank Memorial Fund, New York, pp iv–18
Fenner F, Fantini B (1999) Biological control of vertebrate pests. The history of myxomatosis. An experiment in evolution. CABI, New York
Fleck L (1979) Genesis and development of a scientific fact. Chicago University Press, Chicago [First published 1935]
Frank SA, Schmid-Hempel P (2008) Mechanisms of pathogenesis and the evolution of parasite virulence. J Evol Biol 21:396–404
Fraser C, Hollingsworth TD, Chapman R et al (2007) Variation in HIV-1 set-point viral load: epidemiological analysis and an evolutionary hypothesis. Proc Natl Acad Sci U S A 104(44):17441–17446
Fraser C, Cummings DAT, Klinkenberg D et al (2011) Influenza transmission in households during the 1918 pandemic. Am J Epidemiol 174(5):505–514
Friesen TL, Stukenbrock EH, Lui Z et al (2006) Emergence of a new disease as a result of interspecific virulence gene transfer. Nat Genet 38(8):953–956
Fujimura JH, Chou DY (1994) Dissent in science: styles of scientific practice and the controversy over the cause of AIDS. Soc Sci Med 38(8):1017–1036
Garrett L (1994) The coming plague: newly emerging diseases in a world out of balance. Farrar, Straus and Giroux, New York
Gibbs MJ, Gibbs AJ (2006) Was the 1918 pandemic caused by a bird flu? Nature 440:E8
Godfrey-Smith PJ (2006) The strategy of model-based science. Biol Philos 21:725–740
Goto H, Kawaoka Y (1998) A novel mechanism for the acquisition of virulence by a human influenza A virus. Proc Natl Acad Sci U S A 95:10224–10228
Graham AL, Allen JE, Read A (2005) Evolutionary causes and consequences of immunopathology. Annu Rev Ecol Evol Syst 36:373–397
Grmek MD (1969) Préliminaires d’une étude historique des maladies. Annales ESC 24:1473–1483
Grmek MD (1993) Le concept de maladie émergente. Hist Philos Life Sci 15:281–296
Groisman EA, Ochman H (1996) Pathogenicity islands: bacterial evolution in quantum leaps. Cell 87:791–794
Hacking I (1992) Styles for historians and philosophers. Stud Hist Philos Sci 23(1):1–20
Herfst S, Schrauwen EJA, Linster M, Chutinimitkul S, de Wit E, Munster VJ, Sorrell EM, Bestebroer TM, Burke DF, Smith DJ, Rimmelzwaan GF, Osterhaus ADME, Fouchier RAM (2012) Airborne transmission of influenza A/H5N1 virus between ferrets. Science 336(6088): 1534–1541. doi:10.1126/science.1213362
Holmes EC (2004) 1918 and all that. Science 303(5665):1787–1788
Honigsbaum MA (2013) History of the great influenza pandemic. Death, panic, and hysteria 1830–1920. I.B. Tauris Academic, London
Imai M, Watanabe T, Hatta M, Das SC, Ozawa M, Shinya K, Gongxun Zhong, Hanson A, Katsura H, Watanabe S, Chengjun Li, Kawakami E, Yamada S, Kiso M, Suzuki Y, Maher EA, Neumann G, Kawaoka Y (2012) Experimental adaptation of an influenza H5 HA confers respiratory droplet transmission to a reassortant H5 HA/H1N1 virus in ferrets. Nature. doi:10.1038/nature10831
Jackson RJ, Ramsay AJ, Christensen CD, Beaton S, Hall DF, Ramshaw IA (2001) Expression of mouse interleukin-4 by a recombinant ectromelia virus suppresses cytolytic lymphocyte responses and overcomes genetic resistance to mousepox. J Virol 75:1205–1210
Johnson N (2006) Britain and the 1918–19 Influenza pandemic. A Dark Epilogue. Routledge, New York
Johnson NPAS, Mueller J (2002) Updating the account: global mortality of the 1918–1920 « Spanish » Influenza pandemic. Bull Hist Med 76(1):105–115
Jones EK, Patel NG, Levy MA, Storeygard A, Balk D, Gittleman JL, Daszak P (2008) Global trends in emerging infectious diseases. Nature 451:990–994
Kawaoka Y, Watanabe T (2011) Pathogenesis of the 1918 pandemic influenza virus. PLOS Pathogens 7(1):1–4
Keating P, Cambrosio A (2007) Cancer clinical trials: the emergence and development of a new style of practice. Bull Hist Med 81(1):197–223
Kilbourne ED (ed) (1960) The severity of influenza as a reciprocal of host susceptibility. In virus virulence and pathogenicity. Churchill, London
Kilbourne ED (1977) Influenza pandemics in perspective. JAMA 237(12):1225–1227
King N (2004) The scale politics of emerging diseases. Osiris 19:62–76
Kobasa D, Takada A, Shinya K et al (2004) Enhanced virulence of influenza A viruses with the haemagglutinin of the 1918 pandemic virus. Nature 431:703–707
Kolate G (2005) http://www.nytimes.com/2005/10/06/health/06flu.html?pagewanted=all
Krause RM (1998) Introduction. In: Krause RM (ed) Emerging infections. Biomedical research report. Academic Press, London
Laland KN, Sterelny K, Odling-Smee J, Hoppitt W, Uller T (2011) Cause and effect in biology revisited: Is Mayr’s proximate-ultimate dichotomy still useful? Science 334(6062):1512–1516
Langford C (2005) Did the 1918–19 influenza pandemic originate in China? Popul Dev Rev 31(3):473–505
Lederberg J (1993) Viruses and humankind: intracellular symbiosis and evolutionary competition. In: Morse SS (ed) Emerging viruses. Oxford University Press, New York/Oxford, pp 3–9
Lederberg J, Shope RE, Oaks SC (1992) Emerging infections: microbial threats to health in the United States. National Academy Press, Washington, DC
Levin BR (1996) The evolution and maintenance of virulence in microparasites. Emerg Infect Dis 2(2):93–102
Levin BR, Edén CS (1990) Selection and evolution of virulence in bacteria: an ecumenical excursion and modest suggestion. Parasitology 100:103–115
Levin SA, Pimentel D (1981) Selection for intermediate rates of increase in parasite-host systems. Am Nat 111:3–24
Levins R (1994) The challenge of new disease. In: Wilson ME, Levins R, Spielman A (eds) Disease in evolution. Global changes and emergence of infectious diseases. The New York Academy of Sciences, New York, pp xvii–xix
Lipsitch M, Moxon RE (1997) Virulence and transmissibility of pathogens: what is the relationship? Trends Microbiol 5(10):31–37
Maurelli AT (2006) Black holes, antivirulence genes, and gene inactivation in the evolution of bacterial pathogens. Microbiol Rev 267:1–8
May RM, Anderson RM (1983) Parasite host coevolution. In: Futuyama DJ, Slatkin M (eds) Coevolution. Sinauer, Sunderland, pp 186–206
Mayr E (1961) Cause and effect in biology: kinds of causes, predictability, and teleology are viewed by a practicing biologist. Science 134:1501–1506
McNeill WH (1976) Plagues and people. Basil Blackwell, Oxford
Mendelsohn JA (2002) ‘Like all that lives’: biology, medicine and bacteria in the age of Pasteur and Koch. Hist Philos Life Sci 24(1):3–36
Méthot PO (2012a) Why do parasites harm their hosts? On the origin and legacy of Theobald Smith’s ‘law of declining virulence’ – 1900–1980. Hist Philos Life Sci 34:561–601
Méthot P-O (2012b) Historical epistemology of the concept of virulence: molecular, ecological, and evolutionary aspects of emerging infectious diseases in the 19th and 20th century. PhD thesis, University of Exeter & Université Paris 1 (Panthéon-Sorbonne)
Méthot PO (2012c) Understanding pathogens in the era of next-generation sequencing. J Infect Dev Ctries 6(9):689–691
Méthot P-O. (2014) Science and science policy: regulating ‘select agents’ in the age of synthetic biology. Perspect Sci 23(4) (Forthcoming)
Méthot PO, Alizon S (forthcoming) What is a pathogen? Towards a process view of host-parasite interactions. Virulence
Mills CE, Robins JM, Lipstich M (2004) Transmissibility of the 1918 pandemic influenza. Nature 432:904–906
Mitchell S (2008) Unsimple truths: science, complexity, and policy. University of Chicago Press, Chicago
Morange M (2005) Les secrets du vivant. Contre la pensée unique en biologie. La Découverte, Paris
Morange M (2011) What will result from the interaction between functional and evolutionary biology? Stud Hist Philos Biol Biomed Sci 42:69–74
Morens DM, Taubenberger JK (2009) Understanding influenza backward. JAMA 302(6):679–680
Morens DM, Folkers GK, Fauci AS (2004) The challenge of emerging and re-emerging infectious diseases. Nature 430(6996):242–249
Morens DM, Taubenberger JK, Fauci AS (2009) The persistent legacy of the 1918 influenza virus. N Engl J Med 361(3):225–229
Morens DM, Subbarao K, Taubenberger JK (2012) Engineering H5N1 avian influenza viruses to study human adaptation. Nature 486:335–340
Morse SS (1991) Emerging viruses: defining the rules for viral traffic. Perspect Biol Med 34(3):387–409
Morse SS (1993) Examining the origins of emerging viruses. In: Morse SS (ed) Emerging viruses. Oxford University Press, New York/Oxford
Morse SS (1995) Factors in the emergence of infectious diseases. Emerg Infect Dis 1(1):7–15
Morse SS (2007) Pandemic influenza: studying the lessons of history. Proc Natl Acad Sci 104(18):7313–7314
National Science Advisory Board for Security (NSABB) (2011) Guidance for Enhancing personal reliability and strengthening the culture of responsibility. http://osp.od.nih.gov/sites/default/files/resources/CRWG_Report_final.pdf
Nicolle C (1930) Naissance, vie et mort des maladies infectieuses. Félix Alcan, Paris
Noymer A (2010) Epidemics and time. Influenza and tuberculosis during and after the 1918–1919 pandemic. In: Herring DA, Swedlund AC (eds) Plagues and epidemics. Infected spaces past and present. Berg, Oxford/New York
O’Malley MA (2013) When integration fails: prokaryote phylogeny and the tree of life. Stud Hist Philos Biol Biomed Sci 44:551–562
O’Malley MA, Soyer OS (2012) The roles of integration in molecular systems biology. Stud Hist Philos Biol Biomed Sci 43(1):58–68
Olson DR, Simonsen L, Edelson PJ, Morse SS (2005) Epidemiological evidence of an early wave of the 1918 influenza pandemic in New York City. Proc Natl Acad Sci U S A 102(31):11059–11063
Oxford JS (2001) The so-called great Spanish influenza pandemic of 1918 may have originated in France in 1916. Philos Trans R Soc Lond B Biol Sci 356:1857–1859
Oxford JS, Sefton A, Jackson R, Innes W, Daniels RS, Johnson N (2002) World War I may have allowed the emergence of “Spanish” influenza. Lancet Infect Dis 2:111–114
Oxford JS, Lambkin R, Sefton A, Daniels R, Elliot A, Brown R, Gill D (2005) A hypothesis: the conjunction of soldiers, gas, pigs, ducks, geese and horses in Northern France during the Great War provided the conditions for the emergence of the “Spanish” influenza pandemic of 1918–1919. Vaccines 23:940–945
Palumbi SR (2001) Human as the world’s greatest evolutionary force. Science 293:1786–1790
Pasteur L, Chamberland CE, Roux E (1994 [1881]) De l’atténuation des virus et de leur retour à la virulence. In: Pichot A (ed) Louis Pasteur. Écrits scientifiques et médicaux. Flammarion, Coll. Champs Sciences, Paris, pp 251–258
Phillips H, Killingray D (eds) (2003) The Spanish influenza pandemic of 1918–19. New perspectives, Studies in the social history of medicine. Routledge, London/New York
Poulin R, Combes C (1999) The concept of virulence. Parasitol Today 15(12):474–475
Raberg L, Graham AL, Read AF (2009) Decomposing health: tolerance and resistance to parasites in animals. Philos Trans R Soc 364:37–49
Rappert B (2007) Biotechnology, security and the search for limits. Palgrave Macmillan, New York
Read AF (1994) The evolution of virulence. Trends Microbiol 2(3):73–76
Reid AH et al (2003) The origin of the 1918 pandemic influenza virus: a continuing enigma. J Gen Virol 84:2285–2292
Reid AH, Fanning TG, Hultin JV, Taubenberger JK (1999) Origin and evolution of the 1918 “Spanish” influenza virus hemagglutinin gene. Proc Natl Acad Sci U S A 96:1651–1656
Reid AH, Fanning TG, Janczewski TA, Taubenberger JK (2000) Characterisation of the 1918 “Spanish” Influenza virus neuraminidase gene. Proc Natl Acad Sci U S A 97(12):6785–6790
Reid AH, Janczewski TA, Lourens RM, Elliot AJ, Daniels RS, Berry CL, Oxford JS, Taubenberger JK (2003) Influenza pandemic caused by highly conserved viruses with two receptor-binding variants. Emerg Infect Dis 9(10):1249–1253
Reid AH, Taubenberger JK, Fanning TG (2001) The 1918 Spanish influenza: integrating history and biology. Microbes Infect 3:81–87
Rheinberger HJ (1997) Towards a history of epistemic things. Synthesizing proteins in the test tube. Stanford University Press, Stanford
Rosengard AM, Liu Y, Nie Z, Jimenez R (2002) Variola virus immune evasion design: expression of a highly efficient inhibitor of human complement. Proc Natl Acad Sci U S A 99(13):808–813
Ruse M (1992) Darwinism. In: Keller, Loyds (eds) Keywords in evolutionary biology. Harvard University Press, Cambridge, MA, pp 74–80
Schuenemann VJ et al (2001) Targeted enrichment of ancient pathogens yielding the pPCP1 plasmid of Yersinia pestis from victims of the Black Death. Proc Natl Acad Sci U S A 108(38):E74–E752
Selgelid MJ (2005) Ethics and infectious disease. Bioethics 19(3):272–289
Shaner G, Stromberg EL, Lacy GH, Barker KR, Pirone TP (1992) Nomenclature and concepts of pathogenicity and virulence. Annu Rev Pythopathol 30:47–66
Shapiro-Ilan DI, Fuxa JR, Lacey LA, Onstad DW, Kaya HK (2005) Definitions of pathogenicity and virulence in invertebrate pathology. J Invertebr Pathol 88:1–7
Smith W, Andrewes CH, Laidlaw PP (1933) A virus obtained from influenza patients. Lancet 8:66–68
Snowden FM (2008) Emerging and reemerging diseases: a historical perspective. Immunol Rev 225:9–26
Stevens J, Corper AL, Basler CF, Taubenberger JK, Palese P, Wilson IA (2004) Structure of the uncleaved human H1 hemagglutinin from the extinct 1918 influenza virus. Science 303:1866–1869
Stuart-Harris CH (1953) Influenza. Arnold, London
Taubenberger JF (2005) Chasing the elusive 1918 virus: preparing for the future by examining the past. In: The threat of pandemic influenza: are we ready? Workshop summary, National Academy of Sciences, Washington, DC, pp 69–89
Taubenberger JF (2006) The origin and virulence of the 1918 “Spanish” influenza virus. Proc Am Philos Soc 150:86–112
Taubenberger JK (2003) Genetic characterization of the 1918 ‘Spanish’ influenza pandemic. In: Philips H, Killingray D (eds) The Spanish influenza pandemic of 1918–19. New perspectives. 2003. Studies in the social history of medicine. Routledge, London/New York, pp 39–46
Taubenberger JK, Morens DM (2010) Influenza: the once and future pandemic. Pub Health Rep 125(Supplement 3):16–26
Taubenberger JK, Reid AH, Kraft AE, Bijwaard KE, Fanning TG (1997) Initial genetic characterisation of the 1918 “Spanish” influenza virus. Science 275:1793–1796
Taubenberger JK, Reid AH, Fanning TG (2000) The 1918 influenza virus: a killer comes into view. Virology 274:241–245
Taubenberger JK, Reid AH, Janczewski A, Fanning TG (2001) Integrating historical, clinical and molecular genetic data in order to explain the origin and virulence of the 1918 Spanish influenza virus. Philos Trans R Soc Lond 356:1829–1839
Thomas L (1974) The lives of a cell: notes of biology watcher. Penguin Books, New York
Thomas SR, Elkinton JS (2004) Pathogenicity and virulence. J Invertebr Pathol 85:146–151
Thompson WW, Shay DK, Weintraub E et al (2003) Mortality associated with influenza and respiratory syncytial virus in the United States. JAMA 289(2):179–186
Tumpey T, Garcia-Sastre A, Taubenberger JK, Palese P, Swayne DE, Basler C (2004) Pathogenicity and immunogenicity of influenza viruses with genes from the 1918 pandemic virus. Proc Natl Acad Sci U S A 101:3166–3171
Tumpey TM, Basler CF, Aguilar PV, Zeng H, Solorzano A, Swayne DE, Cox NJ, Katz JM, Taubenberger JK, Palese P, Garcia-Sastre A (2005) Characterization of the reconstructed 1918 Spanish influenza pandemic virus. Science 310(77):77–80
Van Helvoort T (1993) A bacteriological paradigm in influenza research in the first half of the twentieth century. Hist Philos Life Sci 15:3–21
Van Helvoort T (1994) The construction of bacteriophage as bacterial virus: linking endogenous and exogenous thought styles. J Hist Biol 27(1):91–139
Von Bubnoff A (2005) The 1918 flu virus resurrected. Nature. Special report: 794–795
Webby RJ, Webster RG (2003) Are we ready for pandemic influenza? Science 302:1519–1521
Webster RG (1993) Influenza. In: Morse SM (ed) Emerging viruses. Oxford University Press, New York/Oxford, pp 37–45
Webster RG (1999) 1918 Spanish influenza: the secrets remain elusive. Proc Natl Acad Sci U S A 96:1164–1166
Webster RG, Kawaoka Y (1994) Influenza – an emerging and re-emerging disease. Virology 5:103–111
Webster RG, Rott R (1987) Influenza virus A pathogenicity: the pivotal role of hemagglutinin. Cell 50:665–666
Webster RG, Bean WJ, Gorman OT, Chambers TM, Kawaoka Y (1992) Evolution and ecology of influenza A viruses. Microbiol Rev 56(1):152–179
Weir L, Mykhalovski E (2010) Global public health vigilance. Creating a world on alert. Routledge, New York/London
Witte W (2003) The plague that was not allowed to happen. German medicine and the influenza epidemic of 1918–19 in Baden. In: Philips H, Killingray D (eds) The Spanish influenza pandemic of 1918–19. New perspectives. 2003. Studies in the social history of medicine. Routledge, London/New York, 49–57
Wolfe ND, Dunavan CP, Diamond J (2007) Origins of major human infectious diseases. Nature 447:279–283
Acknowledgments
Earlier versions of this article were presented by Pierre-Olivier Méthot at Bristol University as part of the Seminar Series in the Philosophy of Medicine in March 2011 and during the Consortium in the History and Philosophy of Biology at the IHPST, in Paris, in June 2011. Financial support from the SSHRC is gratefully acknowledged (no.752-2007-1257).
Samuel Alizon is funded by an ATIP-avenir grant from CNRS and INSERM.
Author information
Authors and Affiliations
Corresponding author
Editor information
Editors and Affiliations
Rights and permissions
Copyright information
© 2015 Springer Science+Business Media Dordrecht
About this chapter
Cite this chapter
Méthot, PO., Alizon, S. (2015). Emerging Disease and the Evolution of Virulence: The Case of the 1918–1919 Influenza Pandemic. In: Huneman, P., Lambert, G., Silberstein, M. (eds) Classification, Disease and Evidence. History, Philosophy and Theory of the Life Sciences, vol 7. Springer, Dordrecht. https://doi.org/10.1007/978-94-017-8887-8_5
Download citation
DOI: https://doi.org/10.1007/978-94-017-8887-8_5
Published:
Publisher Name: Springer, Dordrecht
Print ISBN: 978-94-017-8886-1
Online ISBN: 978-94-017-8887-8
eBook Packages: Humanities, Social Sciences and LawPhilosophy and Religion (R0)