Abstract
In this report, we compile a study of microbial populations in deep-sea hydrothermal plumes with providing some new data set, and discuss the relationships with geological settings and the type of the hydrothermal system, e.g. ridge or subduction, in the context of the hypothesis “four TAIGAs” (Urabe et al. Chap. 1). Deep-sea hydrothermal plumes represent one of the best habitats for chemolithotrophic microbes to drive primary production in hydrothermal systems. Microbial cell densities in hydrothermal plumes are up to several times more elevated than in the general abyssal seawater. Putative sulfur utilizers, e.g. SUP05 and Alcanivorax in gammaproteobacteria, SAR324 in deltaproteobacteria, and several epsilonproteobacteria, are the dominant microbes that are detected from most of hydrothermal plumes. The microbial community compositions in the plume of an arc-backarc system are different from those of a mid-oceanic ridge hydrothermal system. This is because the cell densities and community composition of the putative sulfur oxidizers may be regulated by reduced sulfur species due to the pH and Eh conditions of the subseafloor and surrounding seawater. Aerobic methanotrophs are found in hydrothermal plumes which contain high concentrations of molecular hydrogen and methane. Quantitative microbial cell analysis by catalyzed reporter deposition based fluorescent in situ hybridization (CARD-FISH) show that the SUP05 populations are 60–100 % responsible for increased microbial cell densities in the hydrothermal plumes of arc-backarc fields. The contribution of the SUP05 cell densities in the plume microbial community is closely connected with the chemical energy from hydrothermal fluids in various types of TAIGA.
You have full access to this open access chapter, Download chapter PDF
Similar content being viewed by others
Keywords
- Deep-sea hydrothermal plume
- Fluorescence in situ hybridization (FISH)
- Quantitative microbial community analysis
- SUP05 (Thioglobus)
1 Introduction to Hydrothermal Plumes and the TAIGA Concept
Deep-sea hydrothermal plumes have been found in hydrothermal fields located in mid-oceanic ridges, subduction zones, and hotspot submarine volcanoes all over the world. Hot hydrothermal fluids emanated from hydrothermal vents are less dense than general abyssal seawater, therefore the fluids rise into the deep sea, mix with surrounding seawater, and form a deep-sea hydrothermal plume. The first, rising stage of hydrothermal plume evolution is termed the buoyant plume. The buoyant plume undergoes substantial dilution with surrounding seawater due to accompanying turbulence. The mixing causes progressive dilution, and then the plume becomes less buoyant among stratified abyssal seawater and evetually reaches some finite maximum height above the seafloor to spread out laterally. This later stage is termed the nonbuoyant plume (or the neutrally buoyant plume) (German and Von Damm 2003).
The hydrothermal fluids are hot and contain many chemical species originated from magma and subseafloor rocks. Therefore the hydrothermal plume, which is formed by the spouting hydrothermal fluid, acts as an important and major interface between the oceanic lithosphere and seawater, and is important for exchanging heat flux and for the reaction between hydrothermal fluid-derived reduced chemicals and oxidants from the surface of the earth. A hydrothermal plume contains both reduced chemicals from hydrothermal fluids and oxidants from seawater, thus a hydrothermal plume is one of the most important habitats for chemolithotrophic microbes in deep-sea hydrothermal systems. According to the TAIGA concept (see Urabe et al. Chap. 1), the chemical components of the hydrothermal fluid and plume are regulated by the geological background, and the hydrothermal systems can be categorized into four groups: (1) sulfur TAIGA with reduced sulfur from magma, (2) methane TAIGA with high methane and ammonium from the sediments around the channels of hydrothermal fluid circulation beneath the seafloor, (3) hydrogen TAIGA with molecular hydrogen (H2) from serpentinization or the breaking of rocks due to fault activity, and (4) iron TAIGA with Fe2+ leached from fresh basalt by acidic hydrothermal fluids. The chemical compositions of the four types of TAIGA control and restrict the microbial community and their functions. In addition, hydrothermal systems are often categorized according to their tectonic background; mid oceanic ridge (MOR) systems, arc-backarc systems, and hot spot systems.
2 Microbial Communities in Hydrothermal Plumes
Chemical signals derived from hydrothermal fluids are widely used to detect hydrothermal plumes as anomalies from the general abyssal seawater. These chemical signals include turbidity, oxidation-reduction potential, 3He, manganese, iron, methane, and molecular hydrogen. Microbial cell densities in a plume are also elevated up to several dozens times higher than those of the general abyssal seawater (Cowen et al. 1986; Naganuma et al. 1989; Winn et al. 1986). This indicates that the microbial community in the plume interacts strongly with the hydrothermal fluid chemicals in the plume. Indeed, the activities of methane oxidation (DeAngelis et al. 1991, 1993; Tsunogai et al. 2000), ammonium oxidation (Lam et al. 2004, 2008), and iron and manganese oxidation (Cowen et al. 1986) have been detected in the plume. Analysis of microbial community structures (Dick and Tebo 2010; Dick et al. 2013; Sunamura et al. 2004) and metagenome/transcriptome (Anantharamana et al. 2013; Dick et al. 2013; Lesniewski et al. 2012) also has shown sulfur oxidation, ammonium oxidation, aerobic methane oxidation, hydrogen oxidation, and heterotrophic activity in the plumes.
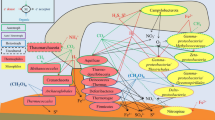
Microbial community structures based on SSU rRNA genes in the plumes have been studied in MOR hydrothermal systems including, for example, Guaymas Basin (Dick and Tebo 2010), Juan de Fuca Ridge (Lam et al. 2008) at the East Pacific Rise (EPR), Mid Cayman (German et al. 2010) at the Mid Cayman Rise, and Kairei Field (Noguchi et al. Chap. 15) at the Central Indian Oceanic Ridge (CIR). These hydrothermal fields are categorized into methane, sulfur, and hydrogen TAIGA fields. In contrast, there has been little study related to microbial communities in arc/backarc hydrothermal systems in subduction region. Only a hydrothermal plume at the Suiyo Seamount (Sunamura et al. 2004) located on the Izu-Ogasawara Arc has been studied. Figure 3.1 shows the microbial community structures in the plume of arc/backarc hydrothermal system, which we collected and analyzed as a representative of methane and sulfur TAIGA. The key microbial lineages in hydrothermal plumes are selected based on a review paper of microbial studies in hydrothermal plumes and anoxic seawater (Dick et al. 2013).
Abundance of key microbial community populations in plumes. The 16S rRNA gene sequences were obtained by cloning/Sanger sequencing for the Daiyon-Yonaguni Knoll, Tarama Knoll, and Wakamiko Creater samples, and by 454 pyrosequencing for the Suiyo Seamount, NW Rota Seamount, TOTO Caldera and CIR samples for this study and Noguchi et al. in Chap. 15. The sequences of Mid Cayman and Guaymus Basin were downloaded from DDBJ or VAMPS. The abbreviated names for hydrothermal zones are as follows: Izu-Ogasawara Arc (IOA), Southern Mariana Arc (SMA), Southern Mariana Trough Backarc (MBA), Southern Okinawa Trough (SOT), Northern Okinawa Trough (NOT), Central Indian Ridge (CIR), Mid Cayman Rise (MCR), and East Pacific Rise (EPR)
In these key microbial linages, the SUP05 phylotype is recognized as the most abundant and universal species in deep-sea hydrothermal plumes of all over the world. The SUP05 phylotype in these plumes was first documented in the Suiyo Seamount hydrothermal plume (Sunamura et al. 2004), and the SUP05 phylotype has been found to be dominant in many anoxic seawater sites (Lavik et al. 2009; Walsh et al. 2009) all over the world. SUP05 is known to be a close relative to symbionts of hydrothermal chemosynthetic mytilids and vesicomyids (Duperron et al. 2005; Fujiwara et al. 2000). These can grow chemolithoautotrophically with sulfur (Marshall and Morris 2012; Walsh et al. 2009) or H2 (Anantharamana et al. 2013; Petersen et al. 2011) as an electron donor, and oxygen or nitrate as an electron acceptor. The representative isolate of SUP05 grow with thiosulfate, which is accumulated as native sulfur vesicles in a cell and, in addition to sulfur compounds, it can grow heterotrophically (Marshall and Morris 2012). This wide variation of energy and carbon metabolism may sustain SUP05 as the primary key species in hydrothermal plume microbial communities. We also found abundant Alcanivorax, SAR324, and several epsilon proteobacterial phylotypes in the plume microbial community. The lineages of these microbes have sulfur-metabolism genes for dissimilar sulfur oxidation (Lai et al. 2012; Nakagawa et al. 2007; Swan et al. 2011) for Alcanivorax, epsilon proteobacteria, and SAR324, respectively. The abundance of potential sulfur oxidizers in the plume is in the good agreement with the result of thermodynamic calculation, which is that sulfur oxidation was the most effective energy source in the mixing zone of deep-sea hydrothermal fluids and general abyssal seawater based on the chemical composition of EPR hydrothermal fluids (McCollom and Shock 1997). On the other hand, 16S rRNA genes of SUP05 lineage was not detected from hydrothermal plume samples in Wakamiko Crater field. In this field, hydrothermal plume shows low pH (6.3–6.8) due to entrainment of volcanic fumaroles in the crater seawater (Yamanaka et al. 2013). The relatively low SUP05 populations was also notable for NW Rota Seamount hydrothermal plume, where very low pH caused by acid sulfate hydrothermal fluid (Resing et al. 2007). In such low pH condition, dominant reduced sulfur species are not HS− but H2S and native sulfur species, and thiosulfate is not stable. In addition, the growth zone of SUP05 lineage is restricted to early stage of hydrothermal plume (see next section), therefore it is essential to import substrates quickly into a cell. Lack of availability of substrates could be responsible for absence of SUP05 in hydrothermal plume. To know the difference in the SUP05 populations between the ridge and arc-backarc hydrothermal systems, further studies such as recycling of SUP05 cells within the plume, activities of dissolved sulfur chemicals, and toxicity of hydrothermal fluid chemicals are needed. Close relatives of SUP05 are able to utilize H2S, HS-, native sulfur, and thiosulfate for growth (Marshall and Morris 2012; Walsh et al. 2009). We proposed here that uptake of the substrate into a cell may be regulated by the stability and states of the sulfur species which caused by pH and Eh.
Figure 3.1 clearly shows that the aerobic methanotrophic lineages are found only in hydrothermal systems where abundant methane and H2 are available (TAIGA of methane and H2). The results indicate that aerobic methanotrophic populations are restricted by high concentrations of methane in hydrothermal fluid as predicted by the TAIGA hypothesis.
Cluster analysis based on the key species populations indicated that the community structures were separated into two groups (Fig. 3.2). The grouping seems to depend on the SUP05 population which is able to use reduced sulfur compounds (Lesniewski et al. 2012; Marshall and Morris 2012) and H2 (Anantharamana et al. 2013; Petersen et al. 2011) as the energy source for chemolithotrophic growth. The first group consists of the plumes of arc-backarc hydrothermal systems with high microbial cell density anomalies. This group contains high SUP05 populations of greater than 50 %. The second group includes the other hydrothermal plumes and could be categorized into three sub-groups. The sub-groups appear to be unrelated to the geological settings of the hydrothermal systems (i.e. ridge or subduction) or to the chemical composition of the original hydrothermal fluids (categories of TAIGA). Analysis of the microbial community structure indicated that the SUP05 lineage was the most important for understanding microbial ecology in the deep-sea hydrothermal plumes.
3 Growth Zone of SUP05
To understand the sulfur and carbon cycles in a hydrothermal field, it is essential to determine the origin and the growth zone of plume-specific microbial communities. Detection and cultivation of hyperthermophiles in an event plume (Summit and Baross 1998) indicates that subseafloor and high temperature hydrothermal fluids are one of the origin of the plume microbes. The SUP05 phylotypes have been detected in the low temperature diffuse hydrothermal flows at a level of more than 99 %(Sunamura et al. 2004), indicating that diffuse flow venting is one of the origins. Quantitative comparison between microbial cell densities and methane concentration among high temperature hydrothermal fluids, low temperature diffuse flows, and hydrothermal plume waters indicated that in situ growth in the plume is most important in the case study of the Suiyo Seamount hydrothermal field (Sunamura et al. 2009) (Fig. 3.3) where SUP05 phylotype dominate microbial communities in the plume (Sunamura et al. 2004). To understand the plume-microbiological impact on deep sea, we need to narrow a more detailed growth zone in the plume.
Variation in ratios of microbial density and hydrothermal chemicals among hydrothermal plumes, low temperature hydrothermal fluids, and high temperature hydrothermal fluids. This figure is cited from Sunamura et al. (2009). Filled circle: Increased in total cell density divided by methane concentration in hydrothermal samples open square: SUP05(typical thiotrophic phylotype in the Suiyo Seamount hydrothermal plume) cell density divided by methane concentration in hydrothermal samples
4 Changes in the Microbial Community During the “Chemical Evolution” of a Plume
Biogeochemical studies of chemical composition of substrates (Malahoff et al. 2006; Resing et al. 2009) and the stable isotope analysis of methane carbon (Tsunogai et al. 2000) in plumes have suggested that the chemical features of a plume change with the dilution process and distance from the vent. This is referred to as the “chemical evolution of plume”. Several previous studies could not find any positive and clear correlation between the total cell density and the physicochemical parameters in the plume. Moreover the changes occurring in the microbial community during the chemical evolution of plume are still unclear. Figure 3.4 shows the relationship between cell densities that are higher than the background (cell density anomalies) and bacteria and SUP05 cell densities determined by the (CARD-)FISH method in various hydrothermal plumes. Good linear correlation between the cell density anomalies and the SUP05 cell densities (Fig. 3.4) indicates that very little “microbial community evolution” occurred during the chemical evolution of the non-buoyant plume.
Relationships between the specific microbial cell population and the total cell population anomaly in three plumes, the Suiyo seamount (data from Sunamura et al. 2004), the Myojin Knoll (this study), and the Haoma Knoll (this study). The total cell density anomalies were estimated by comparison with general abyssal seawater. Red indicates SUP05 cells as determined by a mixture of SUP05-187, 988, and 1245 probes, and green indicates bacterial cells as determined by a mixture of EUB338 I-III probes
5 Contribution of a Specific Microbial Community for Total Plume Microbial Ecosystem
The values of the correlation slope between the microbial cell anomalies and the specific cell densities determined by FISH microscopic counts show the ratios of specific microbial species contributions in the plume microbial communities (Fig. 3.4). Based on these values, bacterial cells are responsible for almost all of the increase in microbial population in the plume and therefore the archaeal contribution is much smaller than that of bacteria. Moreover, SUP05 cells contribute from 60 to 98 % of the anomaly biomass in the plume (Fig. 3.4). Microbial population in the plume is regulated by input and removal; the inputs are in situ chemolithotrophic growth, in situ heterotrophic growth, immigration from vent fluids, and the removals are infection by virus, predation, and sedimentation. Because the contributions of specific microbial populations in the plume as estimated above do not consider removal, the contribution values may be overestimated. Our preliminary result in the southern Mariana plume showed the high correlation coefficient between microbial cell densities and manganese concentration. This indicates that the removal of microbial cells is small compared to the increase in microbial cells. We compared the contribution of SUP05 in the total microbial cell anomaly in the plume with the physical and chemical characteristics of the venting hydrothermal fluid which formed the hydrothermal plume. Within the parameters, we found a close correlation between the SUP05 contribution in the total microbial cell anomaly in the plume and the proportion of sulfur oxidation thermodynamic energy in the expected total thermodynamic energy from mixing of each hydrothermal fluid and abyssal seawater (Fig. 3.5). The positive correlation (r2 = 0.96) suggests that chemical composition of hydrothermal fluids regulates the SUP05 cell density ratios in microbial community, an idea which is proposed in the TAIGA hypothesis. The hydrothermal plume covers and expands from just after hydrothermal fluid spouting out into the seawater to non-buoyant hydrothermal plume. The widespread area and the length of the reaction time in the hydrothermal plume allows most of chemical energy to be used for microbial activity and growth. This idea would explain our conclusion that the composition of hydrothermal fluid chemicals regulates the microbial community structure in the plume. The SUP05 phylotype is known to possess various types of metabolism for growth energy, e.g. sulfur oxidation (Marshall and Morris 2012; Walsh et al. 2009), hydrogen oxidation (Anantharamana et al. 2013; Petersen et al. 2011), and heterotrophy (Marshall and Morris 2012). To understand the regulation mechanism of the plume microbial community, we will need to determine the status of sulfur chemical species, gas species, and metal species during plume evolution in the future.
Relationshop between the SUP05 proportions and the potential chemical energy in plumes of various hydrothermal fields. The SUP05 proportions were determined based on the relationship between SUP05 cell densities and microbial cell density anomalies in the plume from Fig. 3.4 and Sunamura et al., to be submitted. The potential chemical energy is the proportion of potential thermodynamical energy from sulfur oxidation in the total reduced chemical oxidation, which includes methane, reduced sulfur species, dihydrogen (H2), ammonium, iron, and manganese calculated from the end-member chemical concentrations in hydrothermal fluids (Toki et al. Chap. 24, Kawagucci Chap. 35). Thermodynamic calculations were based on (Edwards et al. 2005)
6 Conclusion and Future Perspectives
In this chapter, we compiled our qualitative and quantitative data regarding the plume microbial community of various hydrothermal fields and compare their characteristics inthe context of four TAIGA. We conclude that (1) bacteria is the major player in the hydrothermal plume, (2) SUP05 is the most important key species in the plume microbial community, (3) the SUP05 population represents the arc-backarc and ridge plume communities, and it is regulated by the chemical composition of the hydrothermal fluid, (4) the SUP05 major growth area is restricted to the plumes, not below the seafloor, and (5) methanotrophs are a microbial signature of hydrogen and methane TAIGA. To understand the biogeochemical impacts of plume microbes on the deep ocean, in the future we will need to determine the microbial production rate and in situ microbial activities through in situ incubation, transcriptomics, and more dense sampling.
7 Materials and Methods
7.1 Samples Used in This Study
The hydrothermal plume samples used in this study were collected at the Okinawa Trough (4 sites: Hatoma Knoll in cruise KT05-26, Daiyon-Yonaguni Knoll in KT09-26, Tarama Knoll in NT08-11, and Wakamiko Creater in KT09-29) (see Ishibashi et al. Chap. 29; Yamanaka et al. Chap. 49), and the Izu-Mariana Arc (4sites: Suiyo Seamount in cruise KH11-05, Myojin Caldera in NT06-21, TOTO Caldera in KR08-05, NW Rota Seamount in NT10-13) using a Niskin bottle water sampler with CTD-CMS or ROV Hyper dolphin 3 K. Hydrothermal fluids in the subduction system are rich in sulfides. The hydrothermal fluids in the Okinawa Trough are characterized by a rich methane content. Those in the Ogasawara Arc are characterized by poor content of other reduced chemicals and those of the Mariana Trough, are characterized by a rich iron contents. Microbial cells in the plume samples were filtered and concentrated on 0.22 μm pore sized membrane filters (Type GV, Millipore) for DNA analysis. Other water samples were fixed with neutralized formalin (final concentration: 3.5 %) and microbial cells were filtered on 0.2 μm pore-sized polycarbonate membrane filters (Nuclepore filter, Whatman) on board. These samples were stored at −80 °C prior to analysis.
7.2 Analytical Methods
Microbial DNA on the filter samples was extracted using an Ultra Clean Soil DNA Extraction Kit (MOBio) according to manufacturer’s instructions. To determine the 16S rRNA gene sequences in the samples, we used cloning/Sanger sequencing method or Roche-454 sequencer with a DNA tag for each sample. For the cloning/Sanger method, the 16S rRNA genes were amplified by PCR method with a 27F-1492R primer set (Reysenbach et al. 2000) using Takara ex taq DNA polymerase (Takara bio) and then the amplified 16S rRNA gene sequences were determined by ABI3100 sequencers after cloning using a TA Cloning Kit (Invitrogen). For the Roch-454 sequencer, the 16S rRNA genes were amplified by a 530F-907R primer set (Nunoura et al. 2012) with adaptor and tags by PCR using LA taq DNA polymerase (Takara bio) and the sequences of the amplicons were determined by Roche 454 sequencers. Sequences were analyzed on the Mothur software package (Schloss et al. 2009) using Silva v.1.02. The SUP05 phylotype and bacterial cells were quantified by direct counting under fluorescent microscopy (BX51, Olympus) with CARD-FISH staining of the cells using Cy3-tyramide (Yanagawa and Sunamura in Chap. 6) (Hirayama et al. 2007; Pernthaler et al. 2002) by SUP05 mix probe for the SUP05 phylotype and EUB338 I-III probe for the bacterial cells (Daims et al. 1999; Stahl and Amann 1991). For determination of the SUP05 phylotype, we used a SUP05 mix DNA probe, which is a mixture of newly designed SUP05-988 (5′-AAGTTCCGTGTATGTCAAGA-3′) and SUP05-1245 (5′-GCTTAGCAACCCTTTGTCC-3′) probes, in addition to a SUP05-187 probe (Sunamura et al. 2004).
References
Anantharamana K, Breierb JA, Sheika CS, Dick GJ (2013) Evidence for hydrogen oxidation and metabolic plasticity in widespread deep-sea sulfur-oxidizing bacteria. Proc Natl Acad Sci U S A 110:330–335
Cowen JP, Massoth GJ, Baker ET (1986) Bacterial scavenging of Mn and Fe in a mid-field to far-field hydrothermal particle plume. Nature 322:169–171
Daims H, Bruhl A, Amann R, Schleifer K-H, Wagner M (1999) The domain-specific probe EUB338 is insufficient for the detection of all Bacteria: development and evaluation of a more comprehensive probe set. Syst Appl Microbiol 22:434–440
DeAngelis MA, Baross JA, Lilley MD (1991) Enhanced microbial methane oxidation in water from a deep-sea hydrothermal vent field at simulated in situ hydrostatic pressures. Limnol Oceanogr 36(3):565–570
DeAngelis MA, Lilley MD, Olsen EJ, Baross JA (1993) Methane oxidation in deep-sea hydrothermal plumes of the endeavour segment of the Juan de Fuca Ridge. Deep Sea Res I 40(6):1169–1186
Dick GJ, Tebo BM (2010) Microbial diversity and biogeochemistry of the Guaymas Basin deep-sea hydrothermal plume. Environ Microbiol 12(5):1334–1347
Dick GJ, Anantharaman K, Baker BJ, Li M, Reed DC, Sheik CS (2013) The microbiology of deep-sea hydrothermal vent plumes: ecological and biogeographic linkages to seafloor and water column habitats. Front Microbiol 4:124. doi:10.3389/fmicb.2013.00124
Duperron S, Nadalig T, Caprais JC, Sibuet M, Fiala-Medioni A, Amann R, Dubilier N (2005) Dual symbiosis in a Bathymodiolus sp mussel from a methane seep on the gabon continental margin (southeast Atlantic): 16S rRNA phylogeny and distribution of the symbionts in gills. Appl Environ Microbiol 71(4):1694–1700
Edwards KJ, Bach W, McCollom TM (2005) Geomicrobiology in oceanography: microbe–mineral interactions at and below the seafloor. Trend Microb 13:449–456
Fujiwara Y, Takai K, Uematsu K, Tsuchida S, Hunt JC, Hashimoto J (2000) Phylogenetic characterization of endosymbionts in three hydrothermal vent mussels: influence on host distributions. Mar Ecol Prog Ser 208:147–155
German CR, Von Damm KL (2003) Hydrothermal processes. In: The Oceans and Marine Geochemistry, 1st Edition, H. Elderfield (ed), Treatise on geochemistry, vol 6, Elsevier (Pergamon), Oxford, UK, 625 pp
German CR, Bowen A, Coleman ML, Honig DL, Huber JA, Jakuba MV, Kinsey JC, Kurz MD, Leroy S, McDermott JM, Lépinay BMD, Nakamura K, Seewald JS, Smith JL, Sylva SP, Dover CLV, Whitcomb LL, Yoerger DR (2010) Diverse styles of submarine venting on the ultraslow spreading Mid-Cayman Rise. Proc Natl Acad Sci U S A 107(32):14020–14025
Hirayama H, Sunamura M, Takai K, Nunoura T, Noguchi T, Oida H, Furushima Y, Yamamoto H, Oomori T, Horikoshi K (2007) Culture-dependent and -independent characterization of microbial communities associated with a shallow submarine hydrothermal system occurring within a coral reef off Taketomi Island, Japan. Appl Environ Microbiol 73:7642–7656
Lai Q, Li W, Shao Z (2012) Complete genome sequence of Alcanivorax dieselolei type strain B5. J Bacteriol 194:6674
Lam P, Cowen JP, Jones RD (2004) Autotrophic ammonia oxidation in a deep-sea hydrothermal plume. FEMS Microbiol Ecol 47(2):191–206
Lam P, Cowen JP, Popp BN, Jones RD (2008) Microbial ammonia oxidation and enhanced nitrogen cycling in the endeavour hydrothermal plume. Geochim Cosmochim Acta 72(9):2268–2286
Lavik G, Stuhrmann T, Bruchert V, Van der Plas A, Mohrholz V, Lam P, Mussmann M, Fuchs BM, Amann R, Lass U, Kuypers MMM (2009) Detoxification of sulphidic African shelf waters by blooming chemolithotrophs. Nature 457(7229):581–586
Lesniewski RA, Jain S, Anantharaman K, Schloss PD, Dick GJ (2012) The metatranscriptome of a deep-sea hydrothermal plume is dominated by water column methanotrophs and lithotrophs. ISME J 6:2257–2268
Malahoff A, Kolotyrkina IY, Midson BP, Massoth GJ (2006) A decade of exploring a submarine intraplate volcano: hydrothermal manganese and iron at Lo?’ihi volcano, Hawai’i. Geochem Geophys Geosys 7:Q06002
Marshall KT, Morris RM (2012) Isolation of an aerobic sulfur oxidizer from the SUP05/Arctic96BD-19 clade. ISME J 7:452–455
McCollom TM, Shock EL (1997) Geochemical constraints on chemolithoautotrophic metabolism by microorganisms in seafloor hydrothermal systems. Geochim Cosmochim Acta 61:4375–4391
Naganuma T, Otsuki A, Seki H (1989) Abundance and growth-rate of bacterioplankton community in hydrothermal vent plumes of the North Fiji Basin. Deep Sea Res A 36(9):1379–1390
Nakagawa S, Takaki Y, Shimamura S, Reysenbach A-L, Takai K, Horikoshi K (2007) Deep-sea vent -proteobacterial genomes provide insights into emergence of pathogens. Proc Natl Acad Sci U S A 104:12146–12150
Nunoura T, Takaki Y, Kazama H, Hirai M, Ashi J, Imachi H, Takai K (2012) Microbial diversity in deep-sea methane seep sediments presented by SSU rRNA gene tag sequencing. Microbs Environ 27(4):382–390
Pernthaler A, Pernthaler J, Amann R (2002) Fluorescence in situ hybridization and catalyzed reporter deposition for the identification of marine bacteria. Appl Environ Microbiol 68(6):3094–3101
Petersen JM, Zielinski FU, Pape T, Seifert R, Moraru C, Amann R, Hourdez S, Girguis PR, Wankel SD, Barbe V, Pelletier E, Fink D, Borowski C, Bach W, Dubilier N (2011) Hydrogen is an energy source for hydrothermal vent symbioses. Nature 476:176–180
Resing JA, Lebon G, Baker ET, Lupton JE, Embley RW, Massoth GJ, Chadwick WW, De Ronde CEJ (2007) Venting of acid-sulfate fluids in a high-sulfidation setting at NW rota-1 submarine volcano on the Mariana arc. Econ Geol 102(6):1047–1061
Resing JA, Baker ET, Lupton JE, Walker SL, Butterfield DA, Massoth GJ, Nakamura K (2009) Chemistry of hydrothermal plumes above submarine volcanoes of the Mariana arc. Geochem Geophys Geosyst 10:Q02009
Reysenbach A-L, Longnecker K, Kirshtein J (2000) Novel Bacterial and Archaeal lineages from an in situ growth chamber deployed at a Mid Atlantic Ridge hydrothermal vent. Appl Environ Microbiol 66(9):3798–3806
Schloss PD, Westcott SL, Ryabin T, Hall JR, Hartmann M, Hollister EB, Lesniewski RA, Oakley BB, Parks DH, Robinson CJ, Sahl JW, Stres B, Thallinger GG, Van Horn DJ, Weber CF (2009) Introducing mothur: open-source, platform-independent, community-supported software for describing and comparing microbial communities. Appl Environ Microbiol 75(23):7537–7541
Stahl DA, Amann R (1991) Development and application of nucleic acid probes. In: Stackebrandt E, Goodfellow M (eds) Nucleic acid techniques in bacterial systematics. Wiley, Chichester, pp 205–248
Summit M, Baross JA (1998) Thermophilic subseafloor microorganisms from the 1996 north Gorda Ridge eruption. Deep Sea Res Part II Top Stud Oceanogr 45(12):2751–2766
Sunamura M, Higashi Y, Miyako C, Ishibashi J, Maruyama A (2004) Two Bacteria phylotypes are predominant in the Suiyo Seamount hydrothermal plume. Appl Environ Microbiol 70(2):1190–1198
Sunamura M, Noguchi T, Yamamoto H, Okamura K (2009) Environmental and ecological impact on deep-sea environment from deep-sea hydrothermal system (in Japanese with English abstruct). Chigaku Zasshi 118(6):1160–1173
Swan BK, Martinez-Garcia M, Preston CM, Sczyrba A, Woyke T, Lamy D, Reinthaler T, Poulton NJ, Masland EDP, Gomez ML, Sieracki ME, DeLong EF, Herndl GJ, Stepanauskas R (2011) Potential for chemolithoautotrophy among ubiquitous bacteria lineages in the dark ocean. Science 333:1296–1300
Tsunogai U, Yoshida N, Ishibashi J-I, Gamo T (2000) Carbon isotopic distribution of methane in deep-sea hydrothermal plume, Myojin Knoll Caldera, Izu-Bonin arc: implications for microbial methane oxidation in the oceans and applications to heat flux estimation. Geochim Cosmochim Acta 64:2439–2452
Walsh DA, Zaikova E, Howes CG, Song YC, Wright JJ, Tringe SG, Tortell PD, Hallam SJ (2009) Metagenome of a versatile chemolithoautotroph from expanding oceanic dead zones. Science 326:578–582
Ward JH (1963) Hierarchical grouping to optimize an objective function. J Am Stat Assoc 58:236–244
Winn CD, Karl DM, Massoth GJ (1986) Microorganisms in deep-sea hydrothermal plumes. Nature 320:744–746
Yamanaka T, Maeto K, Akashi H, Ishibashi J, Miyoshi Y, Okamura K, Noguchi T, Kuwahara Y, Toki T, Tsunogai U, Ura T, Nakatani T, Maki T, Kubokawa K, Chiba H (2013) Shallow submarine hydrothermal activity with significant contribution of magmatic water producing talc chimneys in the Wakamiko Crater of Kagoshima Bay, southern Kyushu. Jpn J Volcanol Geotherm Res 258:74–84. doi:10.1016/j.jvolgeores.2013.04.007
Acknowledgements
This work was funded by the Ministry of Education, Culture, Science and Technology (MEXT), Japan, through the Grant-in-Aid for Scientific Research: Project TAIGA (New Scientific Research on Innovative Areas, 20109003) and the Archaean Park Project (International Research Project on Interaction between Sub-Vent Biosphere and Geo-Environments). Thanks are due to the crews of the R/V Tansei-maru, and the R/V Natsushima and the operation teams of HyperDolphin during the KT05-16, KT09-26, NT06-21, NT10-13 cruises. We also thank the on board science parties from all of the cruises and the members of project TAIGA for the sample collection opportunities and for helpful discussions.
Author information
Authors and Affiliations
Corresponding author
Editor information
Editors and Affiliations
Rights and permissions
Open Access This chapter is distributed under the terms of the Creative Commons Attribution Noncommercial License, which permits any noncommercial use, distribution, and reproduction in any medium, provided the original author(s) and source are credited.
Copyright information
© 2015 The Author(s)
About this chapter
Cite this chapter
Sunamura, M., Yanagawa, K. (2015). Microbial Cell Densities, Community Structures, and Growth in the Hydrothermal Plumes of Subduction Hydrothermal Systems. In: Ishibashi, Ji., Okino, K., Sunamura, M. (eds) Subseafloor Biosphere Linked to Hydrothermal Systems. Springer, Tokyo. https://doi.org/10.1007/978-4-431-54865-2_3
Download citation
DOI: https://doi.org/10.1007/978-4-431-54865-2_3
Published:
Publisher Name: Springer, Tokyo
Print ISBN: 978-4-431-54864-5
Online ISBN: 978-4-431-54865-2
eBook Packages: Earth and Environmental ScienceEarth and Environmental Science (R0)