Abstract
Rhythmic heartbeats are paced by electrical impulses that are autonomously generated by cardiac pacemaker cells. This chapter briefly summarizes our recent findings regarding the embryonic origin of and molecular mechanism delineating cardiac pacemaker cells, showing that pacemaker cells are physically segregated and molecularly programmed, in a tertiary heart field, prior to the onset of cardiac morphogenesis.
You have full access to this open access chapter, Download chapter PDF
Similar content being viewed by others
Keywords
- Differentiation
- Fate determination
- Fate mapping
- Lateral mesoderm patterning
- Pacemaker
- Pacemaker cell fate specification
- Tertiary heart field
- Wnt
1 Introduction
Rhythmic heartbeat is initiated by electrical impulses evoked at the sinoatrial node (SAN) that are then conducted to atrial muscle and converge to the atrioventricular node. After a brief delay, pacemaker-initiated action potentials (APs) rapidly pass down the conduction system network and finally spread into ventricular muscle (Fig. 39.1a). The SAN was first described more than a century ago [1], and its anatomical, physiological, and molecular characteristics have been thoroughly investigated [2]. While significant progress has been made in our understanding of the mechanism responsible for the differentiation and patterning of the distal conduction components, the developmental pathway of the SAN remains controversial (Fig. 39.1b).
(a) Diagram of the pacemaking and conduction system network consisting of distinct subcomponents. (b) Controversies in the developmental pathway of SAN pacemaker cells. The classic model assumes that this cell population differentiates from atrial myocytes. A current dogma suggests that SAN pacemaker cells arise from a part of the heart field mesoderm which remains as immature myocytes. These models are based on phenotypic similarities between embryonic myocytes and SAN cells in action potential shape and the expression of unique genes in common. The only way to reliably establish the origins of a cell type is to tag its antecedents. Our direct cell fate mapping studies have identified an origin and novel developmental pathway for SAN pacemaker cells (proposed model)
The identification of the definitive origin of and inductive mechanisms that define SAN pacemaker cells (PCs) will be critical for systematic investigation of developmental mechanisms, such as cell fate specification and differentiation, of this specialized cell population essential for cardiac function. We have recently identified a novel role of Wnt signaling in promoting pacemaker cell induction and differentiation, which is completely contrary to its well-documented inhibitory role in heart field induction [3].
2 Pacemaking Site Transitions From Left To Right During Heart Looping
Classic studies in the chick embryo have shown that as soon as the primitive heart tube forms, myocytes in the posterior inflow tract become electrically active and predominantly evoke pacemaking impulses [4]. While this population has been a priori thought as the progenitor of the SAN pacemaker, no direct cell lineage-tracing study has tested this dogma. Therefore, we have revisited this critical issue. Consistent with previous studies, our optical mapping analysis has detected that the primitive heart tube evokes APs preferentially at the left inflow. Importantly, however, our fate mapping studies have revealed that cells of the left inflow later differentiate into AV junction myocytes rather than the right side pacing cells at the SA [5]. The above findings are consistent with a hypothesis that there are a successive series of pacemaker zones present in the early developing heart [6], indicating that the true origin of SAN pacemaker cells remains to be identified.
3 A Novel Cell Population That Juxtaposes the Right Atrium Takes Over Pacing Function by Mid-heart Looping Stage
While classic and current models assume that SAN PCs arise from a subpopulation of atrial precursors (Fig. 39.1b), to our surprise, our optical mapping data showed that a small region juxtaposing the right atrium preferentially evokes pacemaking action potentials [5]. This pacemaking site has often been neglected in previous studies as this region is routinely dissected away during isolation of the embryonic heart. Our optical mapping and in situ hybridization analyses [5] have identified that this pacemaker site preferentially expresses HCN4 , a major member of the HCN gene family expressed in the heart responsible for the hyperpolarization-activated inward “funny” current, and an atrial-type myosin AMHC1, but is negative for a cardiac transcription factor Nkx2-5 [9]. These are consistent with previous reports on the adult mouse SAN. Other tissues of the conduction system, such as AVN and Purkinje fibers, co-express HCNs and Nkx2-5. Thus, differentiated SAN pacemaker cells can be distinguished by these physiological, pharmacological, and molecular characteristics from other myocytes and conduction cells. Unfortunately, expression of these marker genes is dynamic and no HCN4+/AMHC1+/Nkx2-5- cells can be found in earlier stage embryos. It is therefore unclear when and where SAN pacemaker cell fate is induced and specified. As the heart matures through the processes of looping, heart primordium continually expands with cells being added to both the inflow and outflow segments [7, 8]. The origin of this novel pacemaking cell population needs to be determined.
4 The Right-Sided Pacemaking Cells Indeed Differentiate into SAN Pacemaker Cells
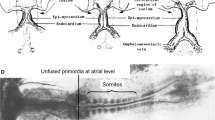
Our in ovo cell-tracing studies have mapped the fate of these extracardiac right-sided pacing cells to the physiologically correct SAN region of the resulting heart at stage 35 (E9) [5]. Optical mapping of these labeled hearts has shown that action potentials are predominantly evoked from these specific cells and propagate into the atrium [5]. Thus, these studies have identified a novel extracardiac cell population as SAN pacemaker precursors. However, until recently it was unknown where this cell population came from. Indeed, the fate of cells in the inflow has been controversial. No systematic fate mapping data was available about the pacemaker field posterior to the heart field at pre-heart tube formation stages. Genetic lineage tracing critically depends upon a gene that is exclusively expressed in pacemaker precursors but not in daughter cells. To our knowledge, no such gene had been identified to date. To explore the origin of SAN pacemaker cells, we performed fate mapping for earlier embryonic stages [5; Fig. 39.2]. For example, in stage 7–8 embryos, the PCs were mapped to a small region in the right lateral plate at somite level 3. Importantly, “the pacemaker field (PF)” does not overlap with “the heart field (HF)” defined by expression of either Nkx2-5 or Isl1 [9, 10]. The data show for the first time that SAN precursors arise from an unpredicted small area of the lateral mesoderm immediately posterior to the known HF. It should be noted that the PF is embedded in the zone that also generates other cardiac-related tissues, such as the proepicardium, right atrium, and vena cava (Bressan 2013). We therefore tentatively termed this previously unrecognized mesodermal area “tertiary heart field,” distinguishing it from the primary heart field and the secondary (anterior) heart field (Fig. 39.3).
Fate map of pacemaker field. (a) At early somite stages, heart precursors occupy bilateral fields within the lateral plate mesoderm. The primary heart field is indicated in yellow, the secondary heart field is indicated in pink, and the posterior tertiary heart field [5] is indicated in blue. Fate mapping studies indicate the progenitors of the SAN pacemaker cells reside within the tertiary heart field (asterisk). (b) At heart tube stages, the primary heart fields have fused along the midline, while the secondary and tertiary heart fields have not yet been incorporated into the heart. The pacemaker precursors maintain their position within the tertiary heart field mesoderm (blue). (c) At looping stages, the pacemaker cells have incorporated into/can be seen attaching the right inflow of the heart and begin to pace the heartbeat. at atria, avj atrioventricular junction, ht heart tube, vt ventricle, a anterior, p posterior, r right, l left
5 Pacemaker Cell Fate Specification Has Already Completed Prior to Heart Morphogenesis
The fate mapping data alone however do not provide any information as to where and when the pacemaker cell fate is specified. A protocol is needed to detect timing and location of cell fate specification apart from marker gene expression. “The cell fate specification” is defined by Harrison [11] and Slack [12] as “when a cell or a tissue becomes capable of autonomously differentiating under a neutral environment, such as in petri dish or test tube.”
Using an established culture model that was previously used for studying classic HF specification, we examined autonomous pacemaker differentiation from PF, which has been mapped in stage 8 embryos (see Fig. 39.2). Under the neutral culture condition, HF explants from stage 8 embryos initiated spontaneous contractions within 24 h. Our data are consistent with previous studies on HF specification. In striking contrast, PF explants did not show any contractions during the first 24 h, but by 48 h many of them started rhythmic contractions with a higher beat rate and exhibited AP waveform characteristic of PCs. PF explants continued to rhythmically beat at a similar rate throughout the extended culture period for at least up to 120 h. The data show that cells posterior to the HF autonomously differentiate to initiate and maintain spontaneous rhythmic contractions, which are distinct from those of the HF.
6 PF Explants Are Sensitive to Blockers Specific for Pacemaking Ion Channel
Our pharmacological tests interrogated whether PF explants use ion channel characteristic of pacemaker cells. Our data show that a HCN channel blocker, ZD7288, induced PF explants to increase the interval between contractions by approximately ~50 % without detectable loss of rhythmicity. An L-type Ca2+ channel blocker, nifedipine, completely diminished spontaneous contraction of PF explants. These pharmacological responses of PF explants are characteristic of SAN pacemaker cells which evoke action potentials mainly through HCN channels and L-type Ca2+ channels and are distinct from cardiomyocytes which mainly use Na+ channels for the upstroke of action potentials.
7 Current Models for Molecular Regulation of SAN Pacemaker Differentiation
It was once postulated that the SAN pacemaker arises from a part of the HF mesoderm which co-expresses Isl1 and Tbx18. Pitx2c , Shox2, Nkx2-5, Tbx3, and Tbx5 are believed to play a role in SAN development. In mice, gene deletion of a laterality gene, Pitx2c, leads to bilateral SANs. Shox2 deficiency results in upregulation of Nkx2-5 and Cx43 in the SAN domain. Both Shox2 and Tbx3 expression require Tbx5. Because Nkx2-5 is absent in the SAN, Nkx2-5 is suggested to suppress HCN4 and Tbx3. Tbx3 gene knockouts result in expression of atrial genes in the SAN and partial loss of SAN-specific gene expression. Ectopic expression of Tbx3 in the atria causes arrhythmia. Taken together, Tbx3 and Tbx18 have been proposed as key transcription factors for SAN pacemaker differentiation. Contradictory to these models, deletion of Tbx3, Tbx18, or Shox2 results in no or only modest pacemaker defects. Further Tbx3 expression occurs at the AV junction and AV node where Nkx2-5 is highly expressed. Our data have revealed, however, that Tbx3, Tbx18, and Isl1 are absent from SAN pacemaker cells during the early stages that correspond with cell fate specification and differentiation. Thus, further elucidation was needed to better understand the molecular mechanisms that regulate the specification and formation of SAN pacemaker cells.
8 A Novel Role of Wnt Signaling for Pacemaker Cell Fate Specification
Heart field induction and specification are promoted by BMP signaling and are restricted to the anterior region by inhibition of Wnt signaling [3, 13]. In chick, Wnt from the neural plate ectoderm and the posterior mesoderm inhibits myocardial differentiation in paraxial and posterior mesoderm, while the secreted Wnt antagonist, crescent, produced by the anterior endoderm supports myocardial development. Noncanonical Wnts that block canonical Wnt/β-catenin signaling can act positively on HF establishment and myocardial electro gradient. Isl1-Cre/β-catenin mutants show defects in outflow tract formation with decreased expression of Tbx2, Tbx3, Shh, and Wnt11. Canonical Wnt signaling is necessary for growth of second HF-derived right ventricular myocytes during heart looping and onward but its role for early specification of the second HF remains obscure. Taken together, inhibition of canonical Wnt/β-catenin signaling is critical for restricting heart field induction to the anterior mesoderm. In contrast to extensive studies of heart field formation, little is known about Wnt signaling in pacemaker development.
Activation of Wnt signaling in the HF diminishes the expression of several cardiac genes, including Nkx2-5. However, it remains largely unknown what HF fate becomes once “HF identity” is lost. Our fate mapping data indicated a surprisingly posterior origin for pacemaker cell progenitors within the Nkx2.5 negative mesoderm, as well as an earlier timing of specification than previously believed. These results led us to reexamine the question of heart field identity following loss of Nkx2.5. Therefore, we investigated this untouched area of Wnt signaling on heart field cells using physiological approaches. Our data have revealed that Wnt-treated heart field cells do not lose contractility either in vivo or in vitro. Instead, they developed a rhythmic, high-rate contraction pattern similar to PF explants, including AP waveforms reminiscent of PCs [5]. A transient exposure to Wnt for only 8 h was sufficient to obtain the pacemaker-like beating pattern. Our in vivo and in vitro data have further demonstrated that inhibition of Wnt signaling for pacemaker progenitors results in a conversion of their fate to the ordinary cardiomyocyte type.
9 Concluding Remarks
An elucidation of the origin and molecular mechanisms that specify the pacemaker cell fate is fundamental to dissecting the earliest steps critical for SAN pacemaker development. Our data have revealed that the pacemaker cell fate is specified in a previously unconsidered embryonic region at very early embryonic stages even before heart morphogenesis begins. The work has also revealed that differential Wnt-mediated signaling cues in the lateral plate mesoderm are sufficient to induce pacemaker-like versus working myocardial fates, and that these fates are maintained throughout early cardiac morphogenesis. These results will significantly increase our understanding of the basis for the mechanisms that regulate pacemaker cell specification and differentiation.
References
Keith A, Flack M. The form and nature of the muscular connections between the primary divisions of the vertebrate heart. J Anat Physiol. 1907;41(Pt 3):172–89.
Boyett MR, Honjo H, Kodama I. The sinoatrial node, a heterogeneous pacemaker structure. Cardiovasc Res. 2000;47(4):658–87.
Marvin MJ, Di Rocco G, Gardiner A, et al. Inhibition of Wnt activity induces heart formation from posterior mesoderm. Genes Dev. 2001;15(3):316–27.
Kamino K, Hirota A, Fujii S. Localization of pacemaking activity in early embryonic heart monitored using voltage-sensitive dye. Nature. 1981;290(5807):595–7.
Bressan M, Liu G, Mikawa T. Early mesodermal cues assign avian cardiac pacemaker fate potential in a tertiary heart field. Science. 2013;340(6133):744–8.
Patten BM. Initiation and early changes in the character of the heart beat in vertebrate embryos. Physiol Rev. 1949;29:31–47.
Jeter Jr JR, Cameron IL. Cell proliferation patterns during cytodifferentiation in embryonic chick tissues: liver, heart and erythrocytes. J Embryol Exp Morphol. 1971;25(3):405–22.
Kirby ML, Gale TF, Stewart DE. Neural crest cells contribute to normal aorticopulmonary septation. Science. 1983;220(4601):1059–61.
Lints TJ, Parsons LM, Hartley L, et al. Nkx-2.5: a novel murine homeobox gene expressed in early heart progenitor cells and their myogenic descendants. Development. 1993;119:419–31.
Cai CL, Liang X, Shi Y, et al. Isl1 identifies a cardiac progenitor population that proliferates prior to differentiation and contributes a majority of cells to the heart. Dev Cell. 2003;5(6):877–89.
Harrison RG. Some difficulties of the determination problem. Am Nat. 1933;67:306–21.
Slack JMW. From egg to embryo: regional specification in early development. New York: Cambridge University Press; 1991.
Schultheiss TM, Burch JB, Lassar AB. A role for bone morphogenetic proteins in the induction of cardiac myogenesis. Genes Dev. 1997;11:451–62.
Acknowledgments
This work has been funded in part by NIH R01078921, R01HL093566, and R01HL112268.
Author information
Authors and Affiliations
Corresponding author
Editor information
Editors and Affiliations
Rights and permissions
Open Access This chapter is distributed under the terms of the Creative Commons Attribution-Noncommercial 2.5 License (http://creativecommons.org/licenses/by-nc/2.5/), which permits any noncommercial use, distribution, and reproduction in any medium, provided the original author(s) and source are credited. The images or other third party material in this chapter are included in the work's Creative Commons license, unless indicated otherwise in the credit line; if such material is not included in the work's Creative Commons license and the respective action is not permitted by statutory regulation, users will need to obtain permission from the license holder to duplicate, adapt or reproduce the material.
Copyright information
© 2016 The Author(s)
About this chapter
Cite this chapter
Bressan, M., Liu, G., Louie, J.D., Mikawa, T. (2016). Cardiac Pacemaker Development from a Tertiary Heart Field. In: Nakanishi, T., Markwald, R., Baldwin, H., Keller, B., Srivastava, D., Yamagishi, H. (eds) Etiology and Morphogenesis of Congenital Heart Disease. Springer, Tokyo. https://doi.org/10.1007/978-4-431-54628-3_39
Download citation
DOI: https://doi.org/10.1007/978-4-431-54628-3_39
Published:
Publisher Name: Springer, Tokyo
Print ISBN: 978-4-431-54627-6
Online ISBN: 978-4-431-54628-3
eBook Packages: MedicineMedicine (R0)