Abstract
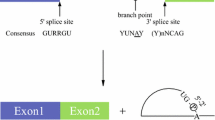
Recent whole genome sequence analyses revealed that a high degree of proteomic complexity is achieved with a limited number of genes. This surprising finding underscores the importance of alternative splicing through which a single gene can generate structurally and functionally distinct protein isoforms [1]. Based on genome-wide analysis, 75% of human genes are thought to encode at least two alternatively spliced isoforms [2, 3]. The regulation of splice site usage provides a versatile mechanism for controlling gene expression and for the generation of proteome diversity, playing essential roles in many biological processes, such as embryonic development, cell growth, and apoptosis. The splice sites are generally recognized by the splicing machinery, a ribonuclear protein complex known as the spliceosome. Spliceosome binding is determined by competing activities of various auxiliary regulatory proteins, such as members of SR protein or heterogeneous nuclear ribonucleoprotein (hnRNP) protein families, which bind specific regulatory sequences and alter the binding of the spliceosome to a particular splice site [1, 4]. Pre-mRNA splicing is regulated in a tissue-specific or developmental stage-specific manner [5]. The selection of splice site can be altered by numerous extracellular stimuli such as hormones, immune response, neuronal depolarization, and cellular stress, through changes in synthesis/degradation, complex formation, and intracellular localization of regulatory proteins. SR proteins are heavily phosphorylated in cells and involved in constitutive and alternative splicing, and the phosphorylation states of SR proteins are altered in response to these extracellular stimuli [6]. Splicing mutations located in either intronic or exonic regions frequently cause hereditary diseases, and more than 15% of mutations that cause genetic disease affect pre-mRNA splicing [7]. Based on a hypothetical idea that we can cure human diseases by regulating the phosphorylation state of SR proteins with synthetic inhibitors of protein kinases, we started our long voyage to challenge the development of new chemical therapeutics.
You have full access to this open access chapter, Download conference paper PDF
Similar content being viewed by others
Keywords
- Splice Site
- Down Syndrome
- Duchenne Muscular Dystrophy
- Severe Acute Respiratory Syndrome
- Duchenne Muscular Dystrophy
These keywords were added by machine and not by the authors. This process is experimental and the keywords may be updated as the learning algorithm improves.
Introduction
Recent whole genome sequence analyses revealed that a high degree of proteomic complexity is achieved with a limited number of genes. This surprising finding underscores the importance of alternative splicing through which a single gene can generate structurally and functionally distinct protein isoforms [1]. Based on genome-wide analysis, 75% of human genes are thought to encode at least two alternatively spliced isoforms [2, 3]. The regulation of splice site usage provides a versatile mechanism for controlling gene expression and for the generation of proteome diversity, playing essential roles in many biological processes, such as embryonic development, cell growth, and apoptosis. The splice sites are generally recognized by the splicing machinery, a ribonuclear protein complex known as the spliceosome. Spliceosome binding is determined by competing activities of various auxiliary regulatory proteins, such as members of SR protein or heterogeneous nuclear ribonucleoprotein (hnRNP) families, which bind specific regulatory sequences and alter the binding of the spliceosome to a particular splice site [1, 4]. Pre-mRNA splicing is regulated in a tissue-specific or developmental stage-specific manner [5]. The selection of splice site can be altered by numerous extracellular stimuli such as hormones, immune response, neuronal depolarization, and cellular stress, through changes in synthesis/degradation, complex formation, and intracellular localization of regulatory proteins. SR proteins are heavily phosphorylated in cells and involved in constitutive and alternative splicing, and the phosphorylation states of SR proteins are altered in response to these extracellular stimuli [6]. Splicing mutations located in either intronic or exonic regions frequently cause hereditary diseases, and more than 15% of mutations that cause genetic disease affect pre-mRNA splicing [7]. Based on a hypothetical idea that we can cure human diseases by regulating the phosphorylation state of SR proteins with synthetic inhibitors of protein kinases, we started our long voyage to challenge the development of new chemical therapeutics.
SRPK Inhibitor SRPIN340 for Viral Diseases and Retinopathy
By extensive screening of 100,000 chemical compounds in a chemical library using in vitro phosphorylation assay, we identified several synthetic chemical compounds that inhibit SR protein kinases (SRPKs) specifically. We found synthetic compounds as specific inhibitors of SRPKs and Cdc-2-like kinases (Clks) and named them SRPIN340 and TG003, respectively [8, 9]. RNA processing plays crucial roles to produce the complexity of viral proteins from the limited size of the gene. A virus utilizes cellular machinery for viral RNA processing, although it is still unclear how virus and cellular machinery interplay and enable achieving the virus propagation. SR proteins have been shown to be involved in the alternative splicing of mRNAs encoding human immunodeficiency virus (HIV)-1 proteins. We found that expression of HIV-1 triggers the dephosphorylation and diminution of SR proteins and that overexpressed SRPK2, one of the SRPKs, preserves the phosphorylation state and protein amounts of SR proteins with the concomitant enhancement of HIV-1 production [8]. These results indicate that SRPKs could be a new target of AIDS therapy. In reality, SRPIN340 suppresses HIV-1 replication in the T-lymphocyte-derived MT-4 cell line and Jurkat cells [8]. SRPIN340 also suppressed propagation of other viruses including Sindbis virus, severe acute respiratory syndrome (SARS) virus, and cytomegalovirus, suggesting that they may require SRPK-dependent SR protein phosphorylation for their multiplication. Recently we found that a derivative of SRPIN340 suppresses virus propagation of the Flaviviridaefamily, which comprises other medically important pathogens including the Japanese encephalitis, yellow fever, and hepatitis C viruses [10]. In addition to SRPK inhibitors, Clk inhibitor TG003 is also reported to suppress propagation of influenza [11].
Vascular endothelial growth factor (VEGF) is a key regulatory component in physiological and pathological angiogenesis. Two types of VEGF isoforms, the pro-angiogenic isoforms and the anti-angiogenic isoforms, are generated by splice site choice in exon 8 [12]. Proximal splice site selection in exon 8 generates pro-angiogenic isoforms such as VEGF165, and distal splice site selection results in anti-angiogenic isoforms such as VEGF165b. Epithelial cells treated with insulin-like growth factor (IGF)-1 increased proximal splice site selection and produced more angiogenic isoforms. In contrast, SRPK1/2 inhibition promoted distal splice site selection and produced more anti-angiogenic isoforms [13]. Injection of SRPIN340 reduced angiogenesis in a mouse model of retinal neovascularization [13]. SRPIN340 or another SRPK inhibitor may be applicable as anticancer drugs by switching the splicing pattern and promoting production of anti-angiogenic VEGF isoforms.
Clk Inhibitor TG003 for Muscular Dystrophy
Recently we found the application of Clk inhibitor TG003 for Duchenne muscular dystrophy (DMD), which is a fatal muscle wasting disease caused by a loss of the dystrophin protein. We met a dystrophinopathy patient who has a point mutation in exon 31 of the dystrophin gene. Although the mutation generates a stop codon, a small amount of internally deleted, but functional, dystrophin protein was produced in the patient’s cells. Analysis of the mRNA revealed that the mutation promotes exon skipping and restores the open reading frame of dystrophin. Presumably, the mutation disrupts an exonic splicing enhancer and creates an exonic splicing silencer. Therefore, we searched for small chemicals that enhance exon skipping, and found that TG003 promoted the skipping of exon 31 in the endogenous dystrophin gene in a dose-dependent manner and increased production of the dystrophin protein in the patient’s cells (Fig. 1) [14]. Although more preclinical studies with animal models are needed, TG003 is the first chemical compound verified to improve dystrophin production in in vitro patient-derived myotubes.
The splicing of Clk1/4 mRNAs is suspended in tissues and cultured cells, and the intermediate forms retaining specific introns are abundantly pooled in the nucleus [15]. Administration of TG003 increased the level of Clk1/4 mature mRNAs by promoting splicing of the intron-retaining RNAs [16]. Under stress conditions, splicing of general pre-mRNAs was inhibited by dephosphorylation of SR splicing factors, but exposure to stresses, such as heat shock and osmotic stress, promoted the maturation of Clk1/4 mRNAs. Clk1/4 proteins translated after heat shock catalyzed rephosphorylation of SR proteins, especially SRSF4 and SRSF10. Our experimental data indicate that Clk1/4 proteins supplied through stress-induced splicing of the intron retaining pre-mRNAs are essential for rapid recovery of the phosphorylation state of SR proteins and contribute to the restart of splicing reactions after stress removal of SR proteins. Therefore, we named ReSCUE (rephosphorylation of SR proteins by Clk urgent expression) for the recovery of SR protein phosphorylation after stress removal [16].
Dyrk Inhibitor INDY for Down Syndrome
Dual-specificity tyrosine-(Y)-phosphorylation-regulated kinase 1A (Dyrk1A) is a serine/threonine kinase that is essential for maintaining the normal development and function of the brain. The physiological importance of Dyrk1A has recently been highlighted by its proposed relationship with the pathogenesis of Down syndrome (DS) [15]. DS is a congenital genetic disorder, caused by a complete or partial trisomy of chromosome 21, and characterized by systemic manifestations including mental retardation, cardiovascular anomalies, craniofacial dysmorphology, malignant neoplasms such as acute myoblastic leukemia, and recurrent infections [17]. Dyrk1A resides within the so-called Down syndrome critical region (DSCR) of human chromosome 21, a genomic region that plays an important role in DS manifestations. The extra copy of the Dyrk1A gene in DS indeed results in a 1.5-fold increase in the Dyrk1A product at both the protein and activity levels in the brain, and the excessive activity is considered a pathogenic factor in Down syndrome [18]. Therefore, we searched for the specific inhibitor of Dyrk1A, and found that a benzothiazole derivative showed a potent inhibitory effect on the kinase activity [19]. X-ray crystallography of the Dyrk1A/INDY complex revealed the binding of INDY in the ATP pocket of the enzyme. INDY effectively reversed the aberrant tau phosphorylation and rescued the repressed NFAT signaling induced by Dyrk1A overexpression. Importantly, proINDY, a prodrug of INDY, effectively recovered Xenopusembryos from head malformation induced by Dyrk1A overexpression, resulting in normally developed embryos and demonstrating the utility of proINDY in vivo [19].
Conclusion
We have developed chemical inhibitors of protein kinases that are involved in splicing regulation. The compounds described here have the possibility to be developed as new therapeutic drugs for neuromuscular diseases, neoplastic diseases, and viral infections by manipulating splicing patterns or transcription level of specific genes.
References
Black DL (2003) Mechanisms of alternative pre-messenger RNA splicing. Annu Rev Biochem 72:291–336
Modrek B, Lee CJ (2003) Alternative splicing in the human, mouse and rat genomes is associated with an increased frequency of exon creation and/or loss. Nat Genet 34:177–180
Johnson JM, Castle J, Garrett-Engele P et al (2003) Genome-wide survey of human alternative pre-mRNA splicing with exon junction microarrays. Science 302:2141–2144
Smith CW, Valcarcel J (2000) Alternative pre-mRNA splicing: the logic of combinatorial control. Trends Biochem Sci 25:381–388
Stamm S (2002) Signals and their transduction pathways regulating alternative splicing: a new dimension of the human genome. Hum Mol Genet 11:2409–2416
Long JC, Caceres JF (2009) The SR protein family of splicing factors: master regulators of gene expression. Biochem J 417:15–27
Krawczak M, Reiss J, Cooper DN (1992) The mutational spectrum of single base-pair substitutions in mRNA splice junctions of human genes: causes and consequences. Hum Genet 90:41–54
Fukuhara T, Hosoya T, Shimizu S, Sumi K, Oshiro T, Yoshinaka Y, Suzuki M, Yamamoto N, Herzenberg LA, Herzenberg LA, Hagiwara M (2006) Utilization of host SR protein kinases and RNA-splicing machinery during viral replication. Proc Natl Acad Sci USA 103:11329–11333
Muraki M, Ohkawara B, Hosoya T, Onogi H, Koizumi J, Koizumi T, Sumi K, Yomoda J, Murray MV, Kimura K, Furuichi K, Shibuya H, Krainer AR, Suzuki M, Hagiwara M (2004) Manipulation of alternative splicing by a newly developed inhibitor of Clks. J Biol Chem 279:24246–24254
Anwar A, Hosoya T, Leong KM, Onogi H, Okuno Y, Hiramatsu T, Koyama H, Suzuki M, Hagiwara M, Garcia-Blanco MA (2011) The Kinase Inhibitor SFV785 Dislocates Dengue Virus Envelope Protein from the Replication Complex and Blocks Virus Assembly. PLoS One 6(8):e23246
Karlas A, Machuy N, Shin Y, Pleissner KP, Artarini A, Heuer D, Becker D, Khalil H, Ogilvie LA, Hess S, Mäurer AP, Müller E, Wolff T, Rudel T, Meyer TF (2010) Genome-wide RNAi screen identifies human host factors crucial for influenza virus replication. Nature (Lond) 463:818–822
Harper SJ, Bates DO (2008) VEGF-A splicing: the key to anti-angiogenic therapeutics? Nat Rev Cancer 8:880–887
Nowak DG, Amin EM, Rennel ES, Hoareau-Aveilla C, Gammons M, Damodoran G, Hagiwara M, Harper SJ, Woolard J, Ladomery MR, Bates DO (2010) Regulation of vascular endothelial growth factor (VEGF) splicing from pro-angiogenic to anti-angiogenic isoforms: a novel therapeutic strategy for angiogenesis. J Biol Chem 285:5532–5540
Nishida A, Kataoka N, Takeshima Y, Yagi M, Awano H, Ota M, Itoh K, Hagiwara M, Matsuo M (2011) Chemical treatment enhances skipping of a mutated exon in the dystrophin gene. Nat Commun 2:308
Duncan PI, Howell BW, Marius RM, Drmanic S, Douville EM, Bell JC (1995) Alternative splicing of STY, a nuclear dual specificity kinase. J Biol Chem 270:21524–21531
Ninomiya K, Kataoka N, Hagiwara M (2011) Stress-responsive maturation of Clk1/4 pre-mRNAs promotes phosphorylation of SR splicing factor. J Cell Biol 195:27–40
Wiseman FK, Alford KA, Tybulewicz VL, Fisher EM (2009) Down syndrome: recent progress and future prospects. Hum Mol Genet 18:R75–R83
Dowjat WK, Adayev T, Kuchna I et al (2007) Trisomy-driven overexpression of DYRK1A kinase in the brain of subjects with Down syndrome. Neurosci Lett 413:77–81
Ogawa Y, Nonaka Y, Goto T, Ohnishi E, Hiramatsu T, Kii I, Yoshida M, Ikura T, Onogi H, Shibuya H, Hosoya T, Ito N, Hagiwara M (2010) Development of a novel selective inhibitor of the Down syndrome-related kinase Dyrk1A. Nat Commun 1:86
Acknowledgments
The author thanks past and present Hagiwara’s laboratory members and collaborators, especially T. Hosoya, A. Nishida, D.G. Nowak, D.O. Bates, M. Matsuo, A. Anwar, M.A. Garcia-Blanco, T. Fukuhara, M. Muraki, N. Kataoka, Y. Ogawa, and H. Onogi, for their contribution to the work described here. This work was supported by Grants-in-Aid from the Ministry of Education, Culture, Sports, Science, and Technology (MEXT) of Japan and research grants from the Japan Science and Technology Agency, Takeda Science Foundation, The Naito Foundation Natural Science Scholarship, and The Uehara Memorial Foundation.
Author information
Authors and Affiliations
Corresponding author
Editor information
Editors and Affiliations
Rights and permissions
Copyright information
© 2012 Springer
About this paper
Cite this paper
Hagiwara, M. (2012). Modulation of Pre-mRNA Splicing Patterns with Synthetic Chemicals and Their Clinical Applications. In: Shibasaki, M., Iino, M., Osada, H. (eds) Chembiomolecular Science. Springer, Tokyo. https://doi.org/10.1007/978-4-431-54038-0_31
Download citation
DOI: https://doi.org/10.1007/978-4-431-54038-0_31
Published:
Publisher Name: Springer, Tokyo
Print ISBN: 978-4-431-54037-3
Online ISBN: 978-4-431-54038-0
eBook Packages: Biomedical and Life SciencesBiomedical and Life Sciences (R0)