Abstract
Sialic acids are components of cell-surface glycans and play important roles in cell–cell communication and host–pathogen interaction. More than 55 genes, encoding receptors, enzymes, and transporters, are known to be involved in sialic acid biology. Nearly 10 years of research have revealed that several of these genes show human-specific changes in genome structure, expression, or function. In this chapter, we introduce these human-specific changes and their possible impact on the human evolution. Also, we give an overview of the evolution of sialic acid biology in primates. The discovery of human-specific changes in sialic acid biology is one step toward explaining the genetic basis of human uniqueness, one of the major activities in primatology, contributing to answering a transdisciplinary question: What makes us human?
You have full access to this open access chapter, Download chapter PDF
Similar content being viewed by others
Keywords
These keywords were added by machine and not by the authors. This process is experimental and the keywords may be updated as the learning algorithm improves.
1 Introduction
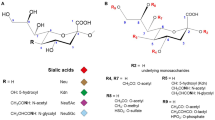
Sialic acids are a family of nine-carbon sugars that are typically found at the terminal end of glycan chains on the cell surface and secreted molecules in the deuterostome lineage of animals (Angata and Varki 2002; Schauer 2000; Varki 2007) (Fig. 8.1a). The two most common forms of sialic acid are N-acetylneuraminic acid (Neu5Ac) and N-glycolylneuraminic acid (Neu5Gc), and Neu5Gc is enzymatically produced from Neu5Ac by adding a single oxygen atom (i.e., hydroxylation) (Fig. 8.1b). A third sialic acid, 2-keto-3 deoxynonulosonic acid (Kdn), is known but is less common in mammals. The amino group can also be rarely remained unmodified (Neu) and is regarded as a fourth sialic acid. These four forms of sialic acids can be further modified in various ways, and this diversity is increased by attachments to the underlying sugar chains by different linkages (α2-3, α2-6, and α2-8) (Angata and Varki 2002; Beyer et al. 1979; Traving and Schauer 1998). This diversity as presented on the cell surface and secreted molecules has been termed the “sialome” (Varki and Angata 2006).
Sialic acids. (a) Sialic acids on cell-surface and secreted molecules. Sialic acids are typically found at the terminal position of glycan chains on the cell surface and secreted molecules. Ac, O-acetyl ester; Fuc, fucose; Gal, galactose; GalNAc, N-acetylgalactosamine; Glc, glucose; GlcNAc, N-acetylglucosamine; Man, mannose; S, sulfate ester; Sia, sialic acid. (b) The two most common forms of sialic acids. The only difference between these two forms is the additional oxygen atom in the N-glycolylgroup of Neu5Gc (arrow). Numbers represent the serial numbers of the nine-carbon backbone. These sialic acids share various α-glycosidic linkages (α2-3, α2-6, and α2-8) to the underlying sugar chain (R) from the C2 position. (Reproduced with permission from Varki 2007)
Sialic acids are synthesized from certain precursor molecules or recycled from glycoconjugates located on the cell surface (Fig. 8.2). They can also be taken up from external sources and metabolically incorporated. The activation of free sialic acid to nucleotide donors (CMP-Neu5Ac, CMP-Neu5Gc, and CMP-Kdn) occurs in the nucleus, and nucleotide donors are then returned to the cytosol for transport into the lumen of the Golgi apparatus. In the Golgi, sialic acid residues are transferred from the CMP donors to newly synthesized glycoconjugates. Finally, sialic acid-containing glycoconjugates are transported to the cell surface, or secreted. Sialic acids on cell surfaces are often recognized by intrinsic and extrinsic molecules and play an important role in cell–cell communication and host–pathogen interaction. The intrinsic recognition is widely involved in many processes including the immune and nervous systems (Varki 2007). Elimination of sialic acid production results in embryonic lethality (Schwarzkopf et al. 2002). On the other hand, several pathogens use sialic acid recognition to initiate their infection (Angata and Varki 2002). More than 55 vertebrate genes are involved in these biochemical and biological processes (Altheide et al. 2006) (Fig. 8.2). A surprisingly large number of these have undergone genomic sequence, expression, or function changes uniquely in the human lineage.
Genes involved in sialic acid biology. All genes or groups of genes thought to be directly involved in sialic acid (Sia) biology are indicated in bold italic font. The question marks represent unknown or hypothetical pathways. AcO, O-acetyl ester; Glc, glucose; GlcNAc, N-acetylglucosamine; Man, mannose; ManNAc, N-acetylmannosamine; STs, sialyltransferases. (Modified from Altheide et al. 2006)
2 Sialic Acid-Related Loci Changed in the Human Lineage
In the first year of the twenty-first century, the draft sequence of human genome was released, heralding a new era of genome biology (International Human Genome Sequencing Consortium 2001). The sequencing of the euchromatic region of the human genome was completed in 2004 (International Human Genome Sequencing Consortium 2004). Following these landmarks, a great deal of effort was made in the genome sequencing projects of non-human primates. This effort resulted in the release of the draft sequences of chimpanzee (Pan troglodytes) and rhesus monkey (Macaca mulatta) in 2005 and 2007, respectively (The Chimpanzee Sequencing and Analysis Consortium 2005; Rhesus Macaque Genome Sequencing and Analysis Consortium 2007). Following these genomic achievements, post-genome biology, in which transcriptome, proteome, and glycome are considered and integrated, has been accelerated in primatology. Such comparative analyses between human and non-human primates are expected to provide us certain hints about the genetic contribution to human uniqueness. Indeed, comparison of human and non-human primate genomes has uncovered the human-specific genomic changes in several genes (Varki and Altheide 2005). Moreover, the transcriptome analysis reveals differences in gene expression between humans and non-human primates (Caceres et al. 2003; Enard et al. 2002; Gilad et al. 2006; Khaitovich et al. 2004a,b, 2005, 2006; Preuss et al. 2004; Somel et al. 2009; Varki and Altheide 2005), as have proteomic and glycomic approaches.
The first example of human-specific change in sialic acid biology was discovered before the advent of genome biology. In 1998, it was found that Neu5Gc, one of the major sialic acids, was completely lacking in humans, but not in our closest evolutionary cousins, the great apes (that is, chimpanzees, bonobos, gorillas, and orangutans) (Muchmore et al. 1998). This complete lack of Neu5Gc was caused by a mutation in the CMAH gene (Chou et al. 1998; Irie et al. 1998) and was a starting point of the voyage to discover human-specific changes in the loci involved in sialic acid biology. At the time of this writing, human-specific changes have been found in 12 loci, equivalent to about 20% of the total known to be involved in sialic acid biology (Varki 2007, 2009): CMAH, SIGLEC1, SIGLEC5, SIGLEC6, SIGLEC7, SIGLEC9, SIGLEC11, SIGLEC12, SIGLEC13, SIGLEC14, SIGLEC16, and ST6GAL1.
The CMAH locus encodes the CMP-N-acetylneuraminic acid hydroxylase (CMAH) enzyme that converts CMP-Neu5Ac to CMP-Neu5Gc in the cytosol (Fig. 8.2), and is essential for producing Neu5Gc (Kawano et al. 1994, 1995). SIGLEC loci encode sialic acid-binding immunoglobulin superfamily lectins (Siglecs), a family of sialic acid recognition proteins (Fig. 8.2). Siglecs are type I transmembrane proteins and mostly expressed in the cells involved in the innate and adaptive immune systems (Crocker 2005; Crocker et al. 2007; Crocker and Varki 2001; Varki and Angata 2006). Their extracellular regions have one V-set immunoglobulin-like domain that binds sialic acids and variable numbers of C2-set immunoglobulin-like domains (Fig. 8.3). An arginine residue in the V-set domain is essential for sialic acid binding (Crocker 2005; Crocker and Varki 2001; Varki and Angata 2006). Many Siglecs have inhibitory signaling motifs (immunoreceptor tyrosine-based inhibitory motif, ITIM) in their cytoplasmic tails (Fig. 8.3), and function as inhibitory receptors in signal transduction of the immune system (Carlin et al. 2009a,b; Crocker 2005; Crocker et al. 2007; Crocker and Varki 2001; Varki and Angata 2006). Other Siglecs lack signaling motifs in their cytoplasmic tails (Fig. 8.3), and some of these can function as activating receptors by the association with adaptor molecules (Angata et al. 2006, 2007; Cao et al. 2008; Crocker et al. 2007). Each of the Siglecs displays a distinct preference for recognizing sialic acid-containing glycans (Crocker et al. 2007; Crocker and Varki 2001; Varki and Angata 2006), an important feature in their function (Carlin et al. 2009b).
Siglec proteins in hominids. Siglecs are type I transmembrane proteins, composed of one V-set immunoglobulin-like domain that binds sialic acids, variable numbers of C2-set immunoglobulin domains, transmembrane domain, and cytoplasmic tail. Many Siglecs have inhibitory motifs (ITIM) in their cytoplasmic tails. On the other hand, Siglecs-14, -15, and -16 have no ITIM in their cytoplasmic tails, but associate with activating adaptor molecules (e.g., DAP12) via charged residues in their transmembrane domains
Siglecs can be divided into two groups: an evolutionarily conserved subgroup (Siglec-1, -2, -4, and -15 in both primates and rodents) and a CD33/Siglec-3-related subgroup [Siglec-3, -5, -6, -7, -8, -9, -10, -11, -12, -13, -14, and -16 in primates; Fig. 8.3; Siglec-3, -E, -F, -G (10), and -H in rodents; Fig. 8.4] (Angata et al. 2004; Crocker et al. 2007; Varki and Angata 2006). The CD33/Siglec-3-related subgroup has experienced changes in gene number in each lineage of eutherian mammals (e.g., 12 genes in the chimpanzee and 5 genes in the mouse) (Angata et al. 2004; Cao et al. 2009; Crocker et al. 2007; Varki and Angata 2006). The ST6GAL1 gene encodes one of the sialyltransferases that transfer sialic acid residues from CMP donor to glycoconjugates in the Golgi and is essential for the production of the α2-6 linkages of sialic acids to N-glycan chains (Martin et al. 2002; Weinstein et al. 1987). Interestingly, all loci that show certain human-specific changes are involved in diversification of sialic acid-containing glycan chains (i.e., sialome) or in sialic acid recognition (see Fig. 8.2). This finding suggests that sialic acid-mediated interactions (e.g., cell–cell communication) have been changed uniquely in the human lineage.
Genomic regions containing CD33/Siglec-3-related Siglec genes in primates and rodents. Siglecs showing an “essential” arginine mutation are indicated by roman numerals (Modified from Angata et al. 2004)
2.1 CMAH
CMAH, the key enzyme that generates variations of sialic acid-containing glycans by producing Neu5Gc (Fig. 8.2), has a Rieske iron–sulfur-binding region as an essential component for enzymatic activity (Schlenzka et al. 1996). Interestingly, the expression of CMAH is specifically suppressed in mammalian brain, which results in the near absence of Neu5Gc in the non-human brain even though Neu5Gc is abundant in other tissues (Gottschalk 1960; Mikami et al. 1998; Muchmore et al. 1998; Nakao et al. 1991; Rosenberg and Schengrund 1976; Schauer 1982; Tettamanti et al. 1965). This is a rather rare case wherein a gene widely expressed in many tissues is selectively downregulated only in brain. The reason why CMAH is downregulated in mammalian brain remains unknown, but some strong selection is likely expected to be behind this unusual situation.
CMAH is a single-copy gene that was inactivated by the deletion of a 92-bp exon (sixth exon) in the human lineage (Chou et al. 1998; Irie et al. 1998; Varki 2001). This deletion, which is found only in humans (Chou et al. 1998; Hayakawa et al. 2001; Irie et al. 1998; Varki 2001), resulted from the insertion of a human-specific Alu element [a SINE (short interspersed element); retroposon] (Hayakawa et al. 2001). As the exon deletion generates a new stop codon in the middle of the gene, humans have only the truncated CMAH protein (Chou et al. 1998). The truncated enzyme lacks the Reiske iron–sulfur cluster region and therefore cannot convert CMP-Neu5Ac to CMP-Neu5Gc (Chou et al. 1998).
Multiple approaches toward estimating the timing of this inactivation (i.e., the extraction of sialic acids from fossils, calculation of pseudogenization time, and estimation of timing of Alu insertion that caused the exon deletion) showed that CMAH was inactivated approximately 3 million years ago (Chou et al. 2002). This calculated timing is compatible with the fact that the CMAH inactivation is universal to modern humans that appeared about 0.2 million years ago (White et al. 2003). Interestingly, the transition from genus Australopithecus to genus Homo occurred about 2–3 million years ago (Carroll 2003; McHenry 1994; Wood 2002; Wood and Collard 1999). Thus, it is possible that transition between these two genera might have involved CMAH inactivation.
One of the major phenotypic differences between the two genera is brain size (McHenry 1994; Wood and Collard 1999). Considering that CMAH is selectively downregulated in the mammalian brain, one possible hypothesis is that the low level of brain Neu5Gc has a suppressive role for brain expansion in other mammals and that CMAH inactivation released our ancestor from this constraint (Chou et al. 2002). However, a Cmah-null mouse did not show any gross increase in brain size (Hedlund et al. 2007). This hypothesis is currently under further evaluation.
Many microbial pathogens initiate their infection by recognizing sialic acids on host cells. Some of them exert a distinct preference for Neu5Gc. For example, enterotoxigenic Escherichia coli K99 adheres to ganglioside GM3 (Neu5Gc), but not to GM (Neu5Ac), in intestinal epithelial cells (Smit et al. 1984). Other pathogens that prefer Neu5Gc for binding include the S40 virus (Campanero-Rhodes et al. 2007), the transmissible gastoenteritis coronavirus (Angata and Varki 2002), and the great ape malaria parasite Plasmodium reichenowi (see below).
As compared with genus Australopithecus, the range of genus Homo expanded widely throughout the Old World, and Homo sapiens to every corner of the globe. After the emergence from Africa, our ancestors undoubtedly met new pathogens in their Out-of-Africa journey. Thus, another hypothesis is that CMAH inactivation protected against the infection by new non-human animal pathogens that prefer Neu5Gc, thereby providing an advantage for the adaptation to a new environment (Hayakawa et al. 2001).
The transgenic mouse that mimics the human inactivation of CMAH, that is, the Cmah-deficient mouse, shows a variety of phenotypes: age-associated decrease in hearing, histological abnormalities in the inner ears, and defects in wound healing (Hedlund et al. 2007). Of these phenotypes, the age-associated decrease in hearing reminds us of the age-associated hearing loss that is common in humans. In addition, abnormality in the inner ears could potentially cause a mild deterioration in balance sense (Hedlund et al. 2007). In this regard, it is interesting that the shift from a mixed arboreal (climbing) and terrestrial (walking) behavior to a primarily terrestrial lifestyle seems to occur in the transition to genus Homo (Bramble and Lieberman 2004; Klein 1999).
The foregoing hypotheses are currently waiting for some supporting evidence from further studies and are based on an assumption that inactivation of CMAH was advantageous for our ancestors to survive. The distant inactivation time (∼3 million years ago) of CMAH makes it difficult to detect positive selection on the inactivated CMAH locus. However, the time back to the most recent common ancestor (TMRCA; 2.9 million years) of all human CMAH haplotypes is very close to the CMAH inactivation time (3.2 million years) with a short duration time, which indicates that inactivated CMAH allele may have fixed quickly in our ancestral population after its emergence (Hayakawa et al. 2006). This point is far from adequate to support a positive selection on the inactivated CMAH locus but it is suggestive.
Under the concept of an “arms race” between hosts and pathogens (“Red Queen” effects) (Hamilton et al. 1990; Van Valen 1974), the human-specific inactivation of CMAH (human-specific loss of Neu5Gc) is expected to result in certain adaptive changes in sialic acid recognition molecules of pathogens. A human malaria parasite, Plasmodium falciparum, uses the sialic acid recognition molecule EBA-175 to bind host sialic acids in their invasion of red blood cells (Baum et al. 2003; Camus and Hadley 1985; Tomita et al. 1978). Interestingly, the sialic acid preference of EBA-175 is different between Plasmodium falciparum and its most closely related chimpanzee parasite, Plasmodium reichenowi: that is, EBA-175 of Plasmodium falciparum prefers Neu5Ac to Neu5Gc, but that of Plasmodium reichenowi prefers Neu5Gc (Martin et al. 2005). This Neu5Ac preference of Plasmodium falciparum may be regarded as an adaptation to the human-specific loss of Neu5Gc. Indeed, recent studies have shown that all extant strains of Plasmodium falciparum are likely derived from a single strain of Plasmodium reichenowi (Rich et al. 2009). In this scenario, human ancestors may have escaped from the common ape malaria by eliminating Neu5Gc, but then later became susceptible to a variant that evolved to now recognize the Neu5Ac-rich red blood cells of humans (Varki and Gagneux 2009).
Shiga toxigenic Escherichia coli, which causes serious gastrointestinal disease in humans, can also secrete a subtilase cytotoxin (SubAB) (Paton et al. 2004). The pentameric B subunit of SubAB, which directs target cell uptake after the binding of surface glycans, has a strong preference for Neu5Gc rather than Neu5Ac, and binds Neu5Gc that is incorporated from the diet into human gut epithelia and kidney vasculature (Byres et al. 2008). This mechanism likely confers susceptibility to the gastrointestinal and systemic toxicities of SubAB in humans (Byres et al. 2008). The CMAH inactivation causes the lack of protective Neu5Gc-containing glycoproteins in serum and other body fluids and may thus make humans kidneys hypersusceptible to the toxin. The susceptibility to SubAB expressing Escherichia coli may be a case where CMAH inactivation increases the risk of infectious disease in humans.
2.2 SIGLEC1
SIGLEC1 belongs to the Siglec gene family and encodes Siglec-1 (sialoadhesin). It is composed of one V-set domain, sixteen C2-set domains, a transmembrane domain, and a short cytoplasmic tail without any signaling motifs (Crocker 2005; Crocker et al. 2007; Crocker and Varki 2001; Hartnell et al. 2001) (Fig. 8.3). As compared with other Siglecs that have at most six C2-set domains, Siglec-1 displays a very long extracellular component (Fig. 8.3). In addition to Siglec-1, there are three known Siglecs having no signaling motif in their cytoplasmic tails: Siglec-14, Siglec-15, and Siglec-16 (Angata et al. 2006, 2007; Cao et al. 2008) (Fig. 8.3). Even though these three Siglecs lack signaling motifs in their cytoplasmic tails, they can function as receptors in signal transduction by the association with an adaptor molecule (Angata et al. 2006, 2007; Cao et al. 2008) (see the “SIGLEC5/SIGLEC14” and “SIGLEC11 and SIGLEC16” sections for details; see Fig. 8.3). In contrast to these Siglecs, Siglec-1 has no such intracellular partner molecules and seems to predominantly act as a simple adhesion molecule without signaling properties. These structural features make Siglec-1 unique among Siglecs.
Siglec-1 is evolutionarily conserved in all mammals examined (Crocker et al. 2007; Crocker and Varki 2001; Varki and Angata 2006). Similar to mouse Siglec-1, human Siglec-1 strongly prefers binding Neu5Ac to Neu5Gc (Brinkman-Van der Linden et al. 2000). However, considering that the loss of Neu5Gc results in the enrichment of Neu5Ac in humans, human Siglec-1 has been exposed to an increase of endogenous ligand density. Siglec-1 is expressed in macrophages (Hartnell et al. 2001), but the expression pattern of human Siglec-1 appears unique. In rats and chimpanzees, Siglec-1 on macrophages is found in both the perifollicular zone and marginal zone of the spleen (Brinkman-Van der Linden et al. 2000). In contrast, human Siglec-1 is found only in perifollicular zone (Brinkman-Van der Linden et al. 2000). Furthermore, almost all CD68+ macrophages in the human spleen are Siglec-1 positive, but only a subpopulation of macrophages expresses Siglec-1 in the chimpanzee spleen (Brinkman-Van der Linden et al. 2000). These human-specific changes may have certain implications in Siglec-1 biology.
Macrophages phagocytose cellular debris and pathogens and play an important role in innate immunity. Siglec-1 on macrophages binds to sialic acids on pathogens such as Trypanosoma cruzi and Neisseria meningitidis and increases the pathogen uptake by macrophages (Jones et al. 2003; Monteiro et al. 2005). If the striking expression of Siglec-1 on almost all macrophages in the spleen means the advanced ability of phagocytosis of human macrophages, human Siglec-1 might be a direct case wherein Siglec evolution improved the ability of protection against certain pathogens. Interestingly, most of the Neu5Ac-expressing pathogens are human specific, and Neu5Gc-synthesizing pathogens have never been recognized (Vimr et al. 2004).
2.3 SIGLEC5/SIGLEC14
SIGLEC5 and SIGLEC14 are members of the CD33/Siglec-3-related subset of Siglec genes and make a primate gene cluster with other members (SIGLEC3, SIGLEC6, SIGLEC7, SIGLEC8, SIGLEC9, SIGLEC10, SIGLEC12, and SIGLEC13) (Angata et al. 2004, 2006; Cao et al. 2009; Varki and Angata 2006) (Figs. 8.3 and 8.4). SIGLEC5 and SIGLEC14 are positioned side-by-side in a tail-to-head orientation on the most telomeric end in the CD33/Siglec-3-related Siglec gene cluster (Fig. 8.4) and are conserved in all primates examined (Angata et al. 2004, 2006; Cao et al. 2009; Varki and Angata 2006).
The SIGLEC5 gene consists of nine exons (Yousef et al. 2002), and the SIGLEC14 is composed of seven exons (Angata et al. 2006). The approximately 1.5-kb region including the first four exons of SIGLEC5 shows homology to that of SIGLEC14, which suggests that gene duplication was involved in the emergence of this Siglec gene pair (Angata et al. 2006). Interestingly, the 5′-end (∼1.3-kb part; designated as A/A′) of the similar region shows extremely high similarity (>99%) between SIGLEC5 and SIGLEC14 in all primate lineages studied (Angata et al. 2006). In contrast, the rest of the similar region (∼0.2 kb; designated as B/B′) shows a lesser identity (78%) (Angata et al. 2006). The high similarity in region A/A′ cannot be explained by the paralogous relationship between two primate genes. The only possible explanation is that recurrent gene conversion events have been occurring in the primate lineages (Angata et al. 2006).
Siglec-5 is composed of one V-set domain, three C2-set domains, a transmembrane domain, and a cytoplasmic tail containing an inhibitory signaling motif (ITIM) (Fig. 8.3), and thus functions as an inhibitory receptor (Crocker 2005; Crocker et al. 2007; Crocker and Varki 2001; Varki and Angata 2006). Siglec-14 is a smaller molecule that consists of one V-set domain, two C2-set domains, a transmembrane domain, and a short cytoplasmic tail with no signaling motif (Angata et al. 2006) (Fig. 8.3). However, Siglec-14 has a positively charged residue (arginine) in its transmembrane domain and associates with activating adaptor molecule DAP12 (TYROBP) via its charged residue (Angata et al. 2006) (Fig. 8.3). Unlike Siglec-5, Siglec-14 therefore functions as an activating receptor. In addition to extreme sequence similarity, sialic acid recognition properties and tissue distribution are very similar between human Siglec-5 and Siglec-14 (Angata et al. 2006; Yamanaka et al. 2009). Taken together, Siglec-5 and Siglec-14 are regarded as “paired receptors” (Angata et al. 2006). The role of paired receptors is elusive, but it is proposed that they are involved in fine-tuning of immune responses via a balance in the ligand binding by activating and inhibitory pair. Because the A/A′ region contains the upstream region and an exon that codes a sialic acid-binding domain (i.e., V-set domain), the gene conversion is the most likely a genomic mechanism that ensures that Siglec-5 and Siglec-14 are the paired receptors.
Interestingly, independent gene conversion between SIGLEC5 and SIGLEC14 (S5–S14 gene conversion) is also found in each of the non-human primates studied, such as chimpanzee, gorilla, orangutan, and baboon, which indicates a high frequency of the S5–S14 gene conversion (Angata et al. 2006). A short (TG)n tract is located between the A/A′ and B/B′ regions (Angata et al. 2006) and is an interesting sequence feature in the terms of the high frequency of the S5–S14 gene conversion. Regardless, the high frequency of S5–S14 gene conversion indicates the necessity of gene conversion in the Siglec-5/Siglec-14 pair as paired receptors. More importantly, the species-specific gene conversion suggests that sialic acid recognition properties and tissue distributions of Siglec-5 and Siglec-14 are similar but become unique in each primate species. In addition, the “essential” arginine residue that confers optimal sialic acid recognition to the sialic acid-binding domain is mutated in the great ape Siglec-5 and Siglec-14 (see Fig. 8.4), which results in the reduction of sialic acid binding in the great ape Siglecs (Angata et al. 2006). It is therefore likely that the biological function of Siglec-5/Siglec-14 pair in the human differs from those in the great apes.
A striking contrast of Siglec-5/Siglec-14 expression has been found between human and chimpanzee T cells. Although the majority of chimpanzee CD4+ T cells express Siglec-5 and/or Siglec-14, CD4+ T cells in humans are mostly negative (Nguyen et al. 2006; Yamanaka et al. 2009). The CD4+ T cells are involved in the pathology of common human diseases such as acquired immunodeficiency syndrome (AIDS), bronchial asthma, rheumatoid arthritis, and type I diabetes. In this regard, human immunodeficiency virus (HIV) progression to AIDS is common in humans but rare in chimpanzees (Novembre et al. 1997; Olson and Varki 2003). Moreover, rheumatoid arthritis, bronchial asthma, and type I diabetes have not been reported in great apes (Olson and Varki 2003; Varki and Altheide 2005). The lack of Siglec-5 and Siglec-14 expression on CD4+ T cells may therefore contribute to T-cell overreactivity in these common human diseases.
A functional deletion of SIGLEC14, which is caused by a fusion between SIGLEC5 and SIGLEC14 loci, has recently been found in human populations from around the world (Yamanaka et al. 2009). As mentioned earlier, the approximately 1.3-kb A/A′ region of human SIGLEC14 represents extreme identity with the corresponding region of human SIGLEC5. The gene fusion caused the lack of region unique to SIGLEC14 (i.e., the fusion boundary is in A/A′ region), resulting in the lack of Siglec-14 expression (Yamanaka et al. 2009) but continued expression of Siglec-5-like protein. The Siglec-14 null individuals are apparently healthy, which shows that the loss of Siglec-14 is not deleterious in healthy human populations (Yamanaka et al. 2009). However, the frequency of the SIGLEC14 null allele is significantly higher in Asians than in Africans and Europeans (Yamanaka et al. 2009), suggesting unknown selective pressures. In this regard, it is interesting that group B Streptococcus, a common cause of sepsis and meningitis in human newborns, binds to Siglec-5 via cell wall-anchored β protein and thereby impairs leukocyte phagocytosis, oxidative burst, and extracellular trap production (Carlin et al. 2009a). This interaction increases bacterial resistance to phagocytosis and killing by human leukocytes and is considered as one of the strategies of microbial innate immune evasion (Carlin et al. 2009a). Siglec-14 may be involved in this interaction as a partner molecule of Siglec-5, and its functional deletion may have certain implication on pathogenic infection.
2.4 SIGLEC6
Human SIGLEC6 is located in the CD33/Siglec-3-related Siglec gene cluster on the long arm of chromosome 19 with orthologues in chimpanzee, baboon, and rhesus monkey (Angata et al. 2004; Cao et al. 2009; Varki and Angata 2006; Yousef et al. 2002) (Fig. 8.4). Siglec-6 is composed of one V-set domain, two C2-set domains, a transmembrane domain, and cytoplasmic tail having an ITIM, and is a structurally typical member of CD33/Siglec-3-related Siglecs (Crocker 2005; Crocker et al. 2007; Crocker and Varki 2001; Patel et al. 1999; Varki and Angata 2006) (Fig. 8.3). However, its expression pattern and binding ability are unusual in humans. Human Siglec-6 is found not only on B cells but also on the trophoblast of the placenta, and binds not only to sialic acids but also to leptin, a non-sialic acid-containing protein (hormone) secreted by adipose tissue and placenta (Patel et al. 1999). The placental expression and leptin binding imply that human Siglec-6 may play an important role in placenta biology.
Interestingly, even though Siglec-6 is expressed on B cells of great apes, its placental expression is unique to humans (Brinkman-Van der Linden et al. 2007). Potential ligands (sialic acid-containing glycoproteins) of Siglec-6 are also expressed in placenta, which indicates that Siglec-6 was recruited to placental expression in the human lineage (Brinkman-Van der Linden et al. 2007). Human pregnancy and parturition are unique as compared with that of other mammals, including non-human primates. Human parturition is prolonged as a result of the negotiation between the larger fetal head and small birth canal in the compensation for the gain of the large brain and erect bipedalism (Lovejoy 2005). Human labor tends to be long in duration, whereas chimpanzees give birth more rapidly (Keeling and Roberts 1972). Siglec-6 expression increases with the onset and progression of labor (Brinkman-Van der Linden et al. 2007). By its role as an inhibitory receptor, Siglec-6, whose expression is recruited in the human placenta, might potentially contribute to the prolongation of the birth process in humans. Siglec-6 also binds to leptin, and they colocalize in the placenta (Brinkman-Van der Linden et al. 2007). The plasma leptin concentration increases during labor (Nuamah et al. 2004), and leptin-deficiency mice represent increase of parturition time (Mounzih et al. 1998). Even though leptin is not a dominant ligand, the leptin–Siglec-6 binding might be important in the prolongation of parturition. Interestingly, recent studies have shown that Siglec-6 expression is further upregulated in the placenta of patients with preeclampsia, a uniquely human disease (Winn et al. 2009).
2.5 SIGLEC7 and SIGLEC9
Siglec-7 and Siglec-9 are members of the CD33/Siglec-3-related Siglec subgroup (Crocker 2005; Crocker et al. 2007; Crocker and Varki 2001; Varki and Angata 2006) (Fig. 8.3). The orthologue of Siglec-7 is not found in rodents, but that of Siglec-9 is probably rodent Siglec-E because of their similar gene location and expression pattern (Angata et al. 2004; Varki and Angata 2006) (see Fig. 8.4). Siglec-9 is therefore considered as an ancient Siglec. Because of the close phylogenetic relationship and identical genomic/domain structure (Angata et al. 2004; Yousef et al. 2002), Siglec-7 might have emerged from Siglec-9 via a gene duplication event in the Siglec expansion in primates. Siglec-7 is dominantly expressed on natural killer (NK) cells and weakly on monocytes (Crocker et al. 2007; Nicoll et al. 1999). On the other hand, the expression level of Siglec-9 is high on monocytes, neutrophils, and conventional dendritic cells, but low on NK cells (Crocker et al. 2007; Zhang et al. 2000). Indeed, although expression intensity is different between Siglec-7 and Siglec-9, both Siglecs are found on the same set of immune cells, which suggests that Siglec-7 and Siglec-9 genes retained the similar set of regulatory elements via gene duplication. Siglec-7 and Siglec-9 can both function as inhibitory receptors in these sets of immune cells because of the presence of ITIM in their cytoplasmic tails (Carlin et al. 2009b; Crocker 2005; Crocker et al. 2007; Crocker and Varki 2001; Nicoll et al. 1999; Varki and Angata 2006; Zhang et al. 2000).
Although human Siglec-9 has nonpreferred binding to Neu5Ac and Neu5Gc, chimpanzee and gorilla Siglec-9 represent a strong preference for Neu5Gc-containing ligands (Sonnenburg et al. 2004). This preference indicates that Siglec-9 has changed its sialic acid preference to bind Neu5Ac in the human lineage after the loss of Neu5Gc. Interestingly, the greatest differences of sequence are found in the V-set domains (i.e., sialic acid-binding domains) of human and great ape Siglec-9 (Sonnenburg et al. 2004). Moreover, the nonsynonymous substitution rate is higher than synonymous substitution rate in the exon encoding the V-set domain in the human lineage (Sonnenburg et al. 2004). These findings indicate that accelerated evolution has occurred in the V-set domain of human Siglec-9. As mentioned in the foregoing “CMAH” section, the loss of Neu5Gc occurred in the human lineage (Muchmore et al. 1998; Varki 2001). The acquisition of Neu5Ac binding in human Siglec-9 is therefore thought to be a consequence of adaptive evolution to the human-specific loss of Neu5Gc. Additionally, Siglec-7 shows similar differences in sequence and sialic acid binding between humans and chimpanzees, as does Siglec-9: multiple amino acid changes in the sialic acid-binding domain, chimpanzee Siglec-7 binding to Neu5Gc, and a human Siglec-7 accommodation of Neu5Ac (Sonnenburg et al. 2004). Similar to Siglec-9, human Siglec-7 may have evolved under an adaptation to the loss of Neu5Gc.
Human Siglec-7 and Siglec-9 seem to present interesting cases of likely molecular coevolution between ligands and endogenous receptors in sialic acid biology. Based on this strong coevolutionary relationship, endogenous sialic acids are considered as functional ligands of Siglec molecules and direct the evolution of sialic acid-binding specificity (Sonnenburg et al. 2004). Human-specific loss of Neu5Gc might have also caused changes in sialic acid-binding specificity of other Siglecs (Sonnenburg et al. 2004).
The alteration of sialic acid preference of human Siglec-7 and Siglec-9 might also be implicated in pathogen infection. Campylobacter jejuni, a human pathogen commonly responsible for gastroenteritis, can express a variety of different sialyloligosaccharides in its lipo-oligosaccharides (Avril et al. 2006; Crocker et al. 2007). Siglec-7 binds to Neu5Acα2-8Neu5Acα2-3Gal in lipo-oligosaccharides on this pathogen and increases pathogen binding to NK cells and monocytes (Avril et al. 2006; Crocker et al. 2007). Campylobacter jejuni might thus be exploiting the Neu5Ac accommodation of human Siglec-7 in its infection. On the other hand, Siglec-9 is dominantly expressed on neutrophils, specialized granulocytes that recognize and kill microorganisms, and binds Neu5Acα2-3Galβ1-4GlcNAc units (Carlin et al. 2009b). Interestingly, the human-specific pathogen group B Streptococcus has the same units on its sialylated capsule and can suppress neutrophil function using its sialic acid-rich capsule to engage human Siglec-9 (Carlin et al. 2009b).
2.6 SIGLEC11 and SIGLEC16
Human SIGLEC11 and SIGLEC16 are members of CD33/Siglec-3-related Siglec genes, but located about 1 Mb centromeric of the CD33/Siglec-3-related Siglec gene cluster (Angata et al. 2002; Crocker 2005; Crocker et al. 2007; Varki and Angata 2006) (Fig. 8.4). They are found in a head-to-head orientation about 9 kb apart (Angata et al. 2002; Cao et al. 2008) (Fig. 8.4). The SIGLEC11 gene is also found in non-human primates such as the great apes and rhesus monkey, and SIGLEC16 is clearly identified at least from the chimpanzee (Angata et al. 2004; Cao et al. 2008; Hayakawa et al. 2005; Varki and Angata 2006) (see Fig. 8.4). On the other hand, mouse, dog, and cow have a single Siglec gene or a Siglec-like pseudogene in the orthologous region of their genomes (Cao et al. 2008). The approximately 3-kb genomic part containing the first eight exons of SIGLEC11 represents sequence similarity to SIGLEC16, which indicates an evolutionary kinship between SIGLEC11 and SIGLEC16 (Angata et al. 2002; Hayakawa et al. 2005). Thus, it has been proposed that SIGLEC11 and SIGLEC16 genes emerged via a gene duplication event in the primate lineage (Angata et al. 2002; Cao et al. 2008).
Interestingly, the approximately 2-kb region (designated A/A′) including the first five exons represents extreme identity (99.3%) between human SIGLEC11 and SIGLEC16, whereas the rest of the similar part (designated B/B′) shows 94.6% identity (Hayakawa et al. 2005). In contrast to human genes, chimpanzee orthologues of SIGLEC11 and SIGLEC16 present no such extreme identity in the similar region (Hayakawa et al. 2005). The phylogenetic trees show that the clustering of human SIGLEC11 and SIGLEC16 is found only in the A/A′ region, not in the B/B′ region (Hayakawa et al. 2005). These findings indicate that gene conversion occurred in the A/A′ region exclusively in the human lineage (Hayakawa et al. 2005). In addition, the genetic distance analysis and the phylogenetic tree constructed by adding bonobo, gorilla, and orangutan SIGLEC11 sequences clearly show that SIGLEC16 converted SIGLEC11 (S16 → S11 gene conversion) only in the human lineage (Hayakawa et al. 2005).
The converted part of human SIGLEC11 contains an exon encoding the sialic acid-binding domain (Hayakawa et al. 2005). Indeed, human Siglec-11 shows a different binding ability from chimpanzee Siglec-11 (Hayakawa et al. 2005). Interestingly, Neu5Gc binding is dramatically reduced in human Siglec-11 (Hayakawa et al. 2005). If chimpanzee Siglec-11 shows the ancestral situation of sialic acid binding, this may again be a consequence of adaptive evolution to the human-specific loss of Neu5Gc, as in the case of Siglec-7/Siglec-9.
In addition to the coding region, the upstream region was involved in the S16 → S11 gene conversion (Hayakawa et al. 2005). In contrast to other CD33/Siglec-3-related Siglec genes, the SIGLEC11 gene shows expression in the human brain (Angata et al. 2002); that is, human brain microglia show a positive staining with anti Siglec-11 antibody, but chimpanzee and orangutan brain microglia do not (Hayakawa et al. 2005). Considering that Siglec-11 binds to sialic acids enriched in the brain, SIGLEC11 seems to have been recruited to brain expression in the human lineage. In this regard, it is also interesting to note that human Siglec-11 binds to oligo sialic acids [(Neu5Acα2-8)2–3], which are enriched in the brain (Hayakawa et al. 2005).
The S16 → S11 gene conversion makes an impact on human Siglec-11 evolution. This gene conversion results in the extreme sequence similarity between the extracellular parts of human Siglec-11 and Siglec-16, which suggests that human Siglec-11 and Siglec-16 have the same sialic acid-binding specificity. Human SIGLEC16 is also expressed in the brain (Cao et al. 2008). It appears that human Siglec-16 functions as an activating receptor by the association with an activating adaptor molecule, DAP12, via a positively charged lysine residue in the transmembrane domain (Cao et al. 2008) (Fig. 8.3). As Siglec-11 is an inhibitory receptor because of the presence of ITIM in the cytoplasmic tail (Angata et al. 2002; Crocker 2005; Crocker et al. 2007; Varki and Angata 2006), the S16 → S11 gene conversion may have assured that Siglec-11 became an inhibitory partner of Siglec-16. In other words, human Siglec-11 and Siglec-16 became human-specific paired receptors via the S16 → S11 gene conversion. The emergence of paired receptors may possibly have contributed to the human brain evolution via alteration of microglial function such as the interaction with neural cells.
In this regard, the cytoplasmic tail of Siglec-11 is known to recruit the protein tyrosine phosphatase SHP-1 (Src homology domain 2-containing phosphatase 1) (Angata et al. 2002). The SHP-1-deficient mice show a marked decrease of the number of microglial cells and a slightly smaller brain size than littermate controls (Wishcamper et al. 2001). It is therefore possible to hypothesize that the Siglec-11 recruitment to brain expression is involved in human brain expansion via association with SHP-1 in microglial cells.
As the human SIGLEC16 sequence released by the human genome project has a 4-bp deletion (4-bpΔ) in exon 2, human SIGLEC16 was originally proposed as a pseudogene (SIGLECP16) (Angata et al. 2002; Hayakawa et al. 2005); this is attributed to the high frequency of the 4-bpΔ allele (null allele) of SIGLEC16 in human populations (e.g., ∼40% in the UK population) (Cao et al. 2008). Because the chimpanzee SIGLEC16 released by the chimpanzee genome project has no 4-bp deletion, it is supposed that the 4-bpΔ allele appeared uniquely in the human lineage. Surprisingly, the same situation, that is, the presence of a null allele of the activating Siglec gene, is also found in SIGLEC14 (see the “SIGLEC5/SIGLEC14” section) (Yamanaka et al. 2009), and both Siglec-16 and Siglec-14 are the activating partners of paired receptors. These findings may provide some hint about the role of Siglec paired receptors.
Siglec-11 has been recently proposed as a molecule related to Alzheimer’s disease, a progressive neurodegenerative disorder (Salminen and Kaarniranta 2009). Alzheimer’s disease is characterized by the continuous increase in the numbers and size of β-amyloid plaques (Salminen and Kaarniranta 2009). In this hypothesis, Siglec-11 induces an antiinflammatory response in microglial cells by recognizing sialylated glycolipids or glycoproteins that bind to β-amyloid plaques, which allow plaques to evade the immune surveillance of microglia (Salminen and Kaarniranta 2009). This is interesting, because microglial expression of Siglec-11 is found only in humans (Angata et al. 2002; Hayakawa et al. 2005), and Alzheimer’s disease is common only in humans (Olson and Varki 2003; Varki and Altheide 2005).
2.7 SIGLEC12
In contrast to other Siglecs, Siglec-12 has two V-set domains (Angata et al. 2001; Yousef et al. 2002) (Fig. 8.3). However, the “essential” arginine residue, which confers the optimal sialic acid recognition, is conserved only in the first V-set domain, indicating that only the first V-set domain functions as a sialic acid-binding domain (Angata et al. 2001). This unique second V-set domain does not define Siglec-12 as an evolutionary lone Siglec because the genomic structure of SIGLEC12 is very similar to that of SIGLEC7 (Angata et al. 2001; Yousef et al. 2002). As the Siglec-7 gene has an exon fossil that corresponds to an exon encoding the second V-set domain of Siglec-12 (Angata et al. 2001), it is suggested that Siglec-12 and Siglec-7 are sibling molecules generated via a gene duplication event; this is also supported by the phylogenetic tree analysis (Angata et al. 2001, 2004). On the other hand, despite the close evolutionary kinship with Siglec-7 expressed on immune cells (NK cells and monocytes), Siglec-12 displays a very different expression pattern, that is, expression on the luminal edge of epithelial cells in organs such as the stomach and tonsils (Angata et al. 2001). It seems that the set of regulatory elements was not conserved in the gene duplication generating Siglec-12 and Siglec-7.
The “essential” arginine residue of Siglec-12 is conserved among the great apes but is changed to a cysteine residue in humans (Angata et al. 2001) (see Fig. 8.4). This “essential” arginine mutation causes the loss of sialic acid binding in human Siglec-12 (Angata et al. 2001). Meanwhile, chimpanzee Siglec-12 binds to both Neu5Ac and Neu5Gc but shows strong preference for Neu5Gc (Angata et al. 2001). Sonnnenburg et al. proposed that the lack of the sialic acid-binding ability in human Siglec-12 reflects a “retirement” caused by the human-specific loss of Neu5Gc (Sonnenburg et al. 2004).
2.8 SIGLEC13
The sequence comparison of the CD33/Siglec-3-related Siglec gene cluster between human, chimpanzee, and baboon shows a few species-specific gene losses (Angata et al. 2004) (Fig. 8.4). In humans, the SIGLEC13 locus is completely deleted (Angata et al. 2004; Varki 2007) (Fig. 8.4). To understand the impact of SIGLEC13 deletion on human evolution, further analysis on sialic acid preference and expression pattern in non-human primates is under way.
2.9 ST6GAL1
Sialic acids have three common linkages to acceptor sugars: α2-3, α2-6, and α2-8 linkages (Angata and Varki 2002; Beyer et al. 1979; Traving and Schauer 1998). A striking contrast in the distribution of α2-6-linked sialic acids is found between humans and great apes (Gagneux et al. 2003). The α2-6-linked sialic acids are abundant in the epithelium lining of the human trachea and lung airways, but not in the epithelial goblet cells that secrete heavily sialylated soluble mucins to the lumen of airways (Gagneux et al. 2003). On the other hand, great apes have no α2-6-linked sialic acids in the former cell types but show an abundance in the latter (Gagneux et al. 2003). These findings clearly indicate that human airway epithelium underwent a concerted bidirectional switch in the expression of α2-6-linked sialic acids, that is, a likely upregulation of ST6GALI in the epithelial lining and downregulation in the goblet cells and secreted mucins.
Human influenza virus A and B are known as pathogens that show strong preference for α2-6-linked sialic acids during their infection and target the human respiratory epithelia (Rogers and Paulson 1983). In contrast, avian and other mammalian influenza viruses prefer α2-3-linked sialic acids in their infection (Webster et al. 1992). It is also reported that chimpanzees show attenuation of human influenza infection (Murphy et al. 1992; Snyder et al. 1986; Subbarao et al. 1995). In humans, α2-6-linked sialic acids are abundant in the target cell surfaces but not in mucins that act as potential soluble decoys for viruses. Thus, the uniquely human distribution of α2-6-linked sialic acids in airway epithelium is considered to correlate to the susceptibility of human influenza viruses.
The upregulation of α2-6-linked sialic acids in the epithelial lining in humans is probably explained by the increase in the common α2-6 linkage of sialic acid to galactose on the N-glycan chain (Gagneux et al. 2003; Martin et al. 2002; Weinstein et al. 1987). This sialic acid structure is primarily produced by a sialyltransferase, ST6Gal-I. It is therefore considered that the unique human distribution of α2-6-linked sialic acids results from the altered spatial regulation of ST6Gal-I expression. However, further work is needed to confirm this hypothesis.
3 Overall Evolution of Sialic Acid Biology in the Primate Lineage
Based on their roles, genes involved in sialic acid biology can be divided into five categories: “biosynthesis”; “activation, transport, and transfer”; “modification”; “recognition”; and “recycling and degradation” (Altheide et al. 2006) (see Fig. 8.2). Biosynthesis genes encode several enzymes involved in the production of sialic acid residues from precursor molecules. Activation, transport, and transfer genes encode many enzymes that activate free sialic acid into the nucleotide donor CMP-sialic acid and transport it into the Golgi in which sialic acid residues are transferred to newly synthesized glycoconjugates from the CMP donors. ST6Gal-I falls into this category. The modification category consists of one known member to date, CMAH. Recognition refers to the loci involved in sialic acid recognition, and the major group of this category is the Siglecs. Recycling and degradation genes encode several molecules that function in the release of sialic acid residues from the glycan chain, the breakdown of sialic acid into acylmannosamines and pyruvate, the removal of 9-O-acetyl esters from sialic acid residues, the lysosomal transport of free sialic acid residues to the cytosol, and the stabilization of the sialidase that releases sialic acid residues.
Evolutionary analysis of these represents an overall acceleration across all categories in primates compared with rodents (Altheide et al. 2006), which suggests that the biological functions based on sialic acid biology have evolved more rapidly in the primate lineage.
3.1 Rapid Evolution of Recognition Molecules
Further comparison among categories shows that the recognition category has evolved more rapidly than the others (Altheide et al. 2006). Notably, Siglecs display a higher rate of evolution than other members in the recognition category (Altheide et al. 2006). Indeed, many human-specific changes occurred in Siglecs, especially, in CD33/Siglec-3-related Siglecs. This finding is consistent with human–chimpanzee genomic comparison, which reveals that CD33/Siglec-3-related Siglecs are one of the fastest evolving groups of genes in the entire genome (The Chimpanzee Sequencing and Analysis Consortium 2005). In addition to the human, non-human primates also display some changes in CD33/Siglec-3-related Siglecs (Fig. 8.5). The SIGLEC7 and SIGLEC12 loci are completely deleted in the baboon (Angata et al. 2004). Moreover, the “essential” arginine residue of Siglec-6 changed to leucine in the baboon, suggesting the loss of optimal sialic acid binding in baboon Siglec-6 (Angata et al. 2004). In addition to baboon Siglec-6, the great apes (chimpanzee, gorilla, and orangutan) show the “essential” arginine changed to histidine or tyrosine in both Siglec-5 and Siglec-14 (Angata et al. 2006). Interestingly, these Siglecs also show human-specific changes and might be most changeable in the primate Siglecs. The CD33/Siglec-3-related Siglec subgroup has been uniquely changed in each primate lineage (see Fig. 8.5).
Even within the components of Siglec molecules, the sialic acid-binding domain is the most rapidly evolving (Altheide et al. 2006). It has been proposed that this high rate of evolution of the sialic acid-binding domains is a secondary “Red Queen” effect (Varki and Angata 2006). Namely, the rapid evolution of sialome is caused by the arms race with sialic acid-binding pathogens (primary “Red Queen” effect), and this rapid sialome evolution then forces the sialic acid-binding domains to evolve quickly to keep up with recognizing the changing sialome as “self” (secondary “Red Queen” effect). Even though sialyltransferase sequences are highly conserved in primates, specific gains and losses of sialic acid expression, for example, human unique changes of the distribution of α2-3- and α2-6-linked sialic acids, are found in each primate species (Altheide et al. 2006; Gagneux et al. 2003). Thus, sialic acid-binding domains may have evolved uniquely in each primate species.
3.2 More Rapid Evolution in the Human Lineage?
The human sialome was changed dramatically by the loss of Neu5Gc (Muchmore et al. 1998; Varki 2001), suggesting that the recognition molecules have experienced more sialome changes in the human than in non-human primates. Considering secondary “Red Queen” effects (Varki and Angata 2006), it is supposed that sialic acid-binding domains of human Siglecs have evolved more rapidly than those of non-human primates. This conjecture is consistent with the highest rate of evolution of the sialic acid-binding domain in the human compared with non-human primates (Altheide et al. 2006) and is supported by the human unique alteration in sialic acid-binding ability shown by many Siglecs (i.e., Siglec-5, Siglec-7, Siglec-9, Siglec-11, Siglec-12, and Siglec-14) (see Fig. 8.5).
The loss of Neu5Gc, that is, the CMAH inactivation, occurred about 3 million years ago (Chou et al. 2002). If the changes of sialic acid-binding ability are a consequence of this event, such changes should have occurred after the loss of Neu5Gc. The timing estimation of genomic event (e.g., gene conversion) involved in the change of sialic acid-binding ability would be helpful to test this hypothesis. At this time, it is supposed that the changes of sialic acid-binding ability in human Siglec-5 and Siglec-14 occurred about 3 million years ago (Angata et al. 2006), which is consistent with the hypothesis. Timing estimations for other Siglecs are currently under way. In addition, the emergence of many human-specific pathogens that express Neu5Ac might have also occurred because of the change in inhibitory human Siglecs toward binding Neu5Ac rather than Neu5Gc (Carlin et al. 2009b).
4 Future Directions and Perspectives
An unusually large number of human-specific changes have been found in genomic loci involved in sialic acid biology (Fig. 8.5), suggesting that certain phenotypes expressed by sialic acid biology have changed uniquely in the human lineage. Based on the biological and biochemical roles of the changed molecules (i.e., CMAH, Siglecs, and ST6Gal-I), it is possible that these human-specific changes have contributed to human uniqueness through the alteration in the interaction between sialome and recognition molecules. Because this interaction is known to be important in the immunity and host–pathogen interaction, a major impact of the detected human-specific changes may have been in these biological systems. Additionally, further findings on these human-specific changes might give some explanation on the background of human unique/common diseases such as Alzheimer’s disease and AIDS (Olson and Varki 2003; Varki and Altheide 2005).
Even though the detected changes are undoubtedly unique in the human, we are still speculating on their significance in human biology. Several hypotheses have been proposed to explain the role of each human-specific change. So far, we have difficulty in testing these hypotheses because human and great apes can never be subjects of genetic manipulation. On the other hand, mice may be too distantly related to primates to use them as models. However, genetic manipulating techniques were recently established for the common marmoset (Callithrix jacchus), a species of New World monkeys (Sasaki et al. 2009).
What makes us human? This is a popular question for mankind, and has been posed by many a researcher. Exploration of human uniqueness in sialic acid biology is one of the scientific approaches to answering this question. Scientists could not directly deal with this question in the twentieth century. However, genome biology, which heralded the first year of the twenty-first century (International Human Genome Sequencing Consortium 2001), has given us many useful tools and hints to consider this question, and post-genome biology will allow for further progress.
Abbreviations
- CMAH:
-
CMP-N-acetylneuraminic acid hydroxylase
- CMP-Kdn:
-
Cytidine monophospho-2-keto-3 deoxynonulosonic acid
- CMP-Neu5Ac:
-
Cytidine monophospho-N-acetylneuraminic acid
- CMP-Neu5Gc:
-
Cytidine monophospho-N-glycolylneuraminic acid
- Gal:
-
Galactose
- GlcNAc:
-
N-Acetylglucosamine
- ITIM:
-
Immunoreceptor tyrosine-based inhibitory motif
- Kdn:
-
2-Keto-3 deoxynonulosonic acid
- Neu5Ac:
-
N-Acetylneuraminic acid
- Neu5Gc:
-
N-Glycolylneuraminic acid
- Siglec:
-
Sialic acid-binding immunoglobulin superfamily lectin
References
Altheide TK, Hayakawa T, Mikkelsen TS, Diaz S, Varki N, Varki A (2006) System-wide genomic and biochemical comparisons of sialic acid biology among primates and rodents: evidence for two modes of rapid evolution. J Biol Chem 281:25689–25702
Angata T, Varki A (2002) Chemical diversity in the sialic acids and related alpha-keto acids: an evolutionary perspective. Chem Rev 102:439–469
Angata T, Varki NM, Varki A (2001) A second uniquely human mutation affecting sialic acid biology. J Biol Chem 276:40282–40287
Angata T, Kerr SC, Greaves DR, Varki NM, Crocker PR, Varki A (2002) Cloning and characterization of human Siglec-11. A recently evolved signaling that can interact with SHP-1 and SHP-2 and is expressed by tissue macrophages, including brain microglia. J Biol Chem 277:24466–24474
Angata T, Margulies EH, Green ED, Varki A (2004) Large-scale sequencing of the CD33-related Siglec gene cluster in five mammalian species reveals rapid evolution by multiple mechanisms. Proc Natl Acad Sci USA 101:13251–13256
Angata T, Hayakawa T, Yamanaka M, Varki A, Nakamura M (2006) Discovery of Siglec-14, a novel sialic acid receptor undergoing concerted evolution with Siglec-5 in primates. FASEB J 20:1964–1973
Angata T, Tabuchi Y, Nakamura K, Nakamura M (2007) Siglec-15: an immune system Siglec conserved throughout vertebrate evolution. Glycobiology 17:838–846
Avril T, Wagner ER, Willison HJ, Crocker PR (2006) Sialic acid-binding immunoglobulin-like lectin 7 mediates selective recognition of sialylated glycans expressed on Campylobacter jejuni lipooligosaccharides. Infect Immun 74:4133–4141
Baum J, Pinder M, Conway DJ (2003) Erythrocyte invasion phenotypes of Plasmodium falciparum in The Gambia. Infect Immun 71:1856–1863
Beyer TA, Rearick JI, Paulson JC, Prieels JP, Sadler JE, Hill RL (1979) Biosynthesis of mammalian glycoproteins. Glycosylation pathways in the synthesis of the nonreducing terminal sequences. J Biol Chem 254:12531–12534
Bramble DM, Lieberman DE (2004) Endurance running and the evolution of Homo. Nature (Lond) 432:345–352
Brinkman-Van der Linden EC, Sjoberg ER, Juneja LR, Crocker PR, Varki N, Varki A (2000) Loss of N-glycolylneuraminic acid in human evolution. Implications for sialic acid recognition by siglecs. J Biol Chem 275:8633–8640
Brinkman-Van der Linden EC, Hurtado-Ziola N, Hayakawa T, Wiggleton L, Benirschke K, Varki A, Varki N (2007) Human-specific expression of Siglec-6 in the placenta. Glycobiology 17:922–931
Byres E, Paton AW, Paton JC, Lofling JC, Smith DF, Wilce MC, Talbot UM, Chong DC, Yu H, Huang S, Chen X, Varki NM, Varki A, Rossjohn J, Beddoe T (2008) Incorporation of a non-human glycan mediates human susceptibility to a bacterial toxin. Nature (Lond) 456:648–652
Caceres M, Lachuer J, Zapala MA, Redmond JC, Kudo L, Geschwind DH, Lockhart DJ, Preuss TM, Barlow C (2003) Elevated gene expression levels distinguish human from non-human primate brains. Proc Natl Acad Sci USA 100:13030–13035
Campanero-Rhodes MA, Smith A, Chai W, Sonnino S, Mauri L, Childs RA, Zhang Y, Ewers H, Helenius A, Imberty A, Feizi T (2007) N-Glycolyl GM1 ganglioside as a receptor for simian virus 40. J Virol 81:12846–12858
Camus D, Hadley TJ (1985) A Plasmodium falciparum antigen that binds to host erythrocytes and merozoites. Science 230:553–556
Cao H, Lakner U, de Bono B, Traherne JA, Trowsdale J, Barrow AD (2008) SIGLEC16 encodes a DAP12-associated receptor expressed in macrophages that evolved from its inhibitory counterpart SIGLEC11 and has functional and non-functional alleles in humans. Eur J Immunol 38:2303–2315
Cao H, de Bono B, Belov K, Wong ES, Trowsdale J, Barrow AD (2009) Comparative genomics indicates the mammalian CD33r Siglec locus evolved by an ancient large-scale inverse duplication and suggests all Siglecs share a common ancestral region. Immunogenetics 61:401–417
Carlin AF, Chang YC, Areschoug T, Lindahl G, Hurtado-Ziola N, King CC, Varki A, Nizet V (2009a) Group B Streptococcus suppression of phagocyte functions by protein-mediated engagement of human Siglec-5. J Exp Med 206:1691–1699
Carlin AF, Uchiyama S, Chang YC, Lewis AL, Nizet V, Varki A (2009b) Molecular mimicry of host sialylated glycans allows a bacterial pathogen to engage neutrophil Siglec-9 and dampen the innate immune response. Blood 113:3333–3336
Carroll SB (2003) Genetics and the making of Homo sapiens. Nature (Lond) 422:849–857
Chou HH, Takematsu H, Diaz S, Iber J, Nickerson E, Wright KL, Muchmore EA, Nelson DL, Warren ST, Varki A (1998) A mutation in human CMP-sialic acid hydroxylase occurred after the Homo–Pan divergence. Proc Natl Acad Sci USA 95:11751–11756
Chou HH, Hayakawa T, Diaz S, Krings M, Indriati E, Leakey M, Paabo S, Satta Y, Takahata N, Varki A (2002) Inactivation of CMP-N-acetylneuraminic acid hydroxylase occurred prior to brain expansion during human evolution. Proc Natl Acad Sci USA 99:11736–11741
Crocker PR (2005) Siglecs in innate immunity. Curr Opin Pharmacol 5:431–437
Crocker PR, Varki A (2001) Siglecs, sialic acids and innate immunity. Trends Immunol 22:337–342
Crocker PR, Paulson JC, Varki A (2007) Siglecs and their roles in the immune system. Nat Rev Immunol 7:255–266
Enard W, Khaitovich P, Klose J, Zollner S, Heissig F, Giavalisco P, Nieselt-Struwe K, Muchmore E, Varki A, Ravid R, Doxiadis GM, Bontrop RE, Paabo S (2002) Intra- and interspecific variation in primate gene expression patterns. Science 296:340–343
Gagneux P, Cheriyan M, Hurtado-Ziola N, van der Linden EC, Anderson D, McClure H, Varki A, Varki NM (2003) Human-specific regulation of alpha 2-6-linked sialic acids. J Biol Chem 278:48245–48250
Gilad Y, Oshlack A, Smyth GK, Speed TP, White KP (2006) Expression profiling in primates reveals a rapid evolution of human transcription factors. Nature (Lond) 440:242–245
Gottschalk A (1960) The chemistry and biology of sialic acids and related substance. Cambridge University Press, Cambridge
Hamilton WD, Axelrod R, Tanese R (1990) Sexual reproduction as an adaptation to resist parasites (a review). Proc Natl Acad Sci USA 87:3566–3573
Hartnell A, Steel J, Turley H, Jones M, Jackson DG, Crocker PR (2001) Characterization of human sialoadhesin, a sialic acid binding receptor expressed by resident and inflammatory macrophage populations. Blood 97:288–296
Hayakawa T, Satta Y, Gagneux P, Varki A, Takahata N (2001) Alu-mediated inactivation of the human CMP-N-acetylneuraminic acid hydroxylase gene. Proc Natl Acad Sci USA 98:11399–11404
Hayakawa T, Angata T, Lewis AL, Mikkelsen TS, Varki NM, Varki A (2005) A human-specific gene in microglia. Science 309:1693
Hayakawa T, Aki I, Varki A, Satta Y, Takahata N (2006) Fixation of the human-specific CMP-N-acetylneuraminic acid hydroxylase pseudogene and implications of haplotype diversity for human evolution. Genetics 172:1139–1146
Hedlund M, Tangvoranuntakul P, Takematsu H, Long JM, Housley GD, Kozutsumi Y, Suzuki A, Wynshaw-Boris A, Ryan AF, Gallo RL, Varki N, Varki A (2007) N-Glycolylneuraminic acid deficiency in mice: implications for human biology and evolution. Mol Cell Biol 27:4340–4346
International Human Genome Sequencing Consortium (2001) Initial sequencing and analysis of the human genome. Nature (Lond) 409:860–921
International Human Genome Sequencing Consortium (2004) Finishing the euchromatic sequence of the human genome. Nature (Lond) 431:931–945
Irie A, Koyama S, Kozutsumi Y, Kawasaki T, Suzuki A (1998) The molecular basis for the absence of N-glycolylneuraminic acid in humans. J Biol Chem 273:15866–15871
Jones C, Virji M, Crocker PR (2003) Recognition of sialylated meningococcal lipopolysaccharide by siglecs expressed on myeloid cells leads to enhanced bacterial uptake. Mol Microbiol 49:1213–1225
Kawano T, Kozutsumi Y, Kawasaki T, Suzuki A (1994) Biosynthesis of N-glycolylneuraminic acid-containing glycoconjugates. Purification and characterization of the key enzyme of the cytidine monophospho-N-acetylneuraminic acid hydroxylation system. J Biol Chem 269:9024–9029
Kawano T, Koyama S, Takematsu H, Kozutsumi Y, Kawasaki H, Kawashima S, Kawasaki T, Suzuki A (1995) Molecular cloning of cytidine monophospho-N-acetylneuraminic acid hydroxylase. Regulation of species- and tissue-specific expression of N-glycolylneuraminic acid. J Biol Chem 270:16458–16463
Keeling MR, Roberts JR (1972) The chimpanzee. In: Bourne GH (ed) Histology, reproduction and restraint, vol 5. Karger, New York, pp 143–150
Khaitovich P, Muetzel B, She X, Lachmann M, Hellmann I, Dietzsch J, Steigele S, Do HH, Weiss G, Enard W, Heissig F, Arendt T, Nieselt-Struwe K, Eichler EE, Paabo S (2004a) Regional patterns of gene expression in human and chimpanzee brains. Genome Res 14:1462–1473
Khaitovich P, Weiss G, Lachmann M, Hellmann I, Enard W, Muetzel B, Wirkner U, Ansorge W, Paabo S (2004b) A neutral model of transcriptome evolution. PLoS Biol 2:E132
Khaitovich P, Hellmann I, Enard W, Nowick K, Leinweber M, Franz H, Weiss G, Lachmann M, Paabo S (2005) Parallel patterns of evolution in the genomes and transcriptomes of humans and chimpanzees. Science 309:1850–1854
Khaitovich P, Enard W, Lachmann M, Paabo S (2006) Evolution of primate gene expression. Nat Rev Genet 7:693–702
Klein RG (1999) The human career: human biology and cultural origins. The University of Chicago, Chicago
Lovejoy CO (2005) The natural history of human gait and posture. Part 1. Spine and pelvis. Gait Posture 21:95–112
Martin LT, Marth JD, Varki A, Varki NM (2002) Genetically altered mice with different sialyltransferase deficiencies show tissue-specific alterations in sialylation and sialic acid 9-O-acetylation. J Biol Chem 277:32930–32938
Martin MJ, Rayner JC, Gagneux P, Barnwell JW, Varki A (2005) Evolution of human–chimpanzee differences in malaria susceptibility: relationship to human genetic loss of N-glycolylneuraminic acid. Proc Natl Acad Sci USA 102:12819–12824
McHenry HM (1994) Tempo and mode in human evolution. Proc Natl Acad Sci USA 91:6780–6786
Mikami T, Kashiwagi M, Tsuchihashi K, Daino T, Akino T, Gasa S (1998) Further characterization of equine brain gangliosides: the presence of GM3 having N-glycolyl neuraminic acid in the central nervous system. J Biochem 123:487–491
Monteiro VG, Lobato CS, Silva AR, Medina DV, de Oliveira MA, Seabra SH, de Souza W, DaMatta RA (2005) Increased association of Trypanosoma cruzi with sialoadhesin-positive mice macrophages. Parasitol Res 97:380–385
Mounzih K, Qiu J, Ewart-Toland A, Chehab FF (1998) Leptin is not necessary for gestation and parturition but regulates maternal nutrition via a leptin resistance state. Endocrinology 139:5259–5262
Muchmore EA, Diaz S, Varki A (1998) A structural difference between the cell surfaces of humans and the great apes. Am J Phys Anthropol 107:187–198
Murphy BR, Hall SL, Crowe J, Collins P, Subbarao K, Connors M, London WT, Chanock R (1992) In: Corwin J, Landon JC (eds) Chimpanzee conservation and public health. Diagnon/Bioqual, Rockville, MD, pp 21–27
Nakao T, Kon K, Ando S, Hirabayashi Y (1991) A NeuGc-containing trisialoganglioside of bovine brain. Biochim Biophys Acta 1086:305–309
Nguyen DH, Hurtado-Ziola N, Gagneux P, Varki A (2006) Loss of Siglec expression on T lymphocytes during human evolution. Proc Natl Acad Sci USA 103:7765–7770
Nicoll G, Ni J, Liu D, Klenerman P, Munday J, Dubock S, Mattei MG, Crocker PR (1999) Identification and characterization of a novel siglec, siglec-7, expressed by human natural killer cells and monocytes. J Biol Chem 274:34089–34095
Novembre FJ, Saucier M, Anderson DC, Klumpp SA, O’Neil SP, Brown CR 2nd, Hart CE, Guenthner PC, Swenson RB, McClure HM (1997) Development of AIDS in a chimpanzee infected with human immunodeficiency virus type 1. J Virol 71:4086–4091
Nuamah MA, Yura S, Sagawa N, Itoh H, Mise H, Korita D, Kakui K, Takemura M, Ogawa Y, Nakao K, Fujii S (2004) Significant increase in maternal plasma leptin concentration in induced delivery: a possible contribution of pro-inflammatory cytokines to placental leptin secretion. Endocr J 51:177–187
Olson MV, Varki A (2003) Sequencing the chimpanzee genome: insights into human evolution and disease. Nat Rev Genet 4:20–28
Patel N, Brinkman-Van der Linden EC, Altmann SW, Gish K, Balasubramanian S, Timans JC, Peterson D, Bell MP, Bazan JF, Varki A, Kastelein RA (1999) OB-BP1/Siglec-6: a leptin- and sialic acid-binding protein of the immunoglobulin superfamily. J Biol Chem 274:22729–22738
Paton AW, Srimanote P, Talbot UM, Wang H, Paton JC (2004) A new family of potent AB(5) cytotoxins produced by Shiga toxigenic Escherichia coli. J Exp Med 200:35–46
Preuss TM, Caceres M, Oldham MC, Geschwind DH (2004) Human brain evolution: insights from microarrays. Nat Rev Genet 5:850–860
Rhesus Macaque Genome Sequencing and Analysis Consortium (2007) Evolutionary and biomedical insights from the rhesus macaque genome. Science 316:222–234
Rich SM, Leendertz FH, Xu G, Lebreton M, Djoko CF, Aminake MN, Takang EE, Diffo JL, Pike BL, Rosenthal BM, Formenty P, Boesch C, Ayala FJ, Wolfe ND (2009) The origin of malignant malaria. Proc Natl Acad Sci USA 106(35):14902–14907
Rogers GN, Paulson JC (1983) Receptor determinants of human and animal influenza virus isolates: differences in receptor specificity of the H3 hemagglutinin based on species of origin. Virology 127:361–373
Rosenberg A, Schengrund C (1976) Biological roles of sialic acids. Plenum, New York and London
Salminen A, Kaarniranta K (2009) Siglec receptors and hiding plaques in Alzheimer’s disease. J Mol Med 87:697–701
Sasaki E, Suemizu H, Shimada A, Hanazawa K, Oiwa R, Kamioka M, Tomioka I, Sotomaru Y, Hirakawa R, Eto T, Shiozawa S, Maeda T, Ito M, Ito R, Kito C, Yagihashi C, Kawai K, Miyoshi H, Tanioka Y, Tamaoki N, Habu S, Okano H, Nomura T (2009) Generation of transgenic non-human primates with germline transmission. Nature (Lond) 459:523–527
Schauer R (1982) Sialic acids: chemistry, metabolism and function. Springer, New York
Schauer R (2000) Achievements and challenges of sialic acid research. Glycoconj J 17:485–499
Schlenzka W, Shaw L, Kelm S, Schmidt CL, Bill E, Trautwein AX, Lottspeich F, Schauer R (1996) CMP-N-acetylneuraminic acid hydroxylase: the first cytosolic Rieske iron–sulphur protein to be described in Eukarya. FEBS Lett 385:197–200
Schwarzkopf M, Knobeloch KP, Rohde E, Hinderlich S, Wiechens N, Lucka L, Horak I, Reutter W, Horstkorte R (2002) Sialylation is essential for early development in mice. Proc Natl Acad Sci USA 99:5267–5270
Smit H, Gaastra W, Kamerling JP, Vliegenthart JF, de Graaf FK (1984) Isolation and structural characterization of the equine erythrocyte receptor for enterotoxigenic Escherichia coli K99 fimbrial adhesin. Infect Immun 46:578–584
Snyder MH, London WT, Tierney EL, Maassab HF, Murphy BR (1986) Restricted replication of a cold-adapted reassortant influenza A virus in the lower respiratory tract of chimpanzees. J Infect Dis 154:370–371
Somel M, Franz H, Yan Z, Lorenc A, Guo S, Giger T, Kelso J, Nickel B, Dannemann M, Bahn S, Webster MJ, Weickert CS, Lachmann M, Paabo S, Khaitovich P (2009) Transcriptional neoteny in the human brain. Proc Natl Acad Sci USA 106:5743–5748
Sonnenburg JL, Altheide TK, Varki A (2004) A uniquely human consequence of domain-specific functional adaptation in a sialic acid-binding receptor. Glycobiology 14:339–346
Subbarao K, Webster RG, Kawaoka Y, Murphy BR (1995) Are there alternative avian influenza viruses for generation of stable attenuated avian-human influenza A reassortant viruses? Virus Res 39:105–118
Tettamanti G, Bertona L, Berra B, Zambotti V (1965) Glycolyl-neuraminic acid in ox brain gangliosides. Nature (Lond) 206:192
The Chimpanzee Sequencing and Analysis Consortium (2005) Initial sequence of the chimpanzee genome and comparison with the human genome. Nature (Lond) 437:69–87
Tomita M, Furthmayr H, Marchesi VT (1978) Primary structure of human erythrocyte glycophorin A. Isolation and characterization of peptides and complete amino acid sequence. Biochemistry 17:4756–4770
Traving C, Schauer R (1998) Structure, function and metabolism of sialic acids. Cell Mol Life Sci 54:1330–1349
Van Valen L (1974) Two modes of evolution. Nature (Lond) 252:298–300
Varki A (2001) Loss of N-glycolylneuraminic acid in humans: mechanisms, consequences, and implications for hominid evolution. Am J Phys Anthropol Suppl 33:54–69
Varki A (2007) Glycan-based interactions involving vertebrate sialic-acid-recognizing proteins. Nature (Lond) 446:1023–1029
Varki A (2009) Multiple changes in sialic acid biology during human evolution. Glycoconj J 26:231–245
Varki A, Altheide TK (2005) Comparing the human and chimpanzee genomes: searching for needles in a haystack. Genome Res 15:1746–1758
Varki A, Angata T (2006) Siglecs: the major subfamily of I-type lectins. Glycobiology 16:1R–27R
Varki A, Gagneux P (2009) Human-specific evolution of sialic acid targets: explaining the malignant malaria mystery? Proc Natl Acad Sci USA 106:14739–14740
Vimr ER, Kalivoda KA, Deszo EL, Steenbergen SM (2004) Diversity of microbial sialic acid metabolism. Microbiol Mol Biol Rev 68:132–153
Webster RG, Bean WJ, Gorman OT, Chambers TM, Kawaoka Y (1992) Evolution and ecology of influenza A viruses. Microbiol Rev 56:152–179
Weinstein J, Lee EU, McEntee K, Lai PH, Paulson JC (1987) Primary structure of beta-galactoside alpha 2,6-sialyltransferase. Conversion of membrane-bound enzyme to soluble forms by cleavage of the NH2-terminal signal anchor. J Biol Chem 262:17735–17743
White TD, Asfaw B, DeGusta D, Gilbert H, Richards GD, Suwa G, Howell FC (2003) Pleistocene Homo sapiens from Middle Awash, Ethiopia. Nature (Lond) 423:742–747
Winn VD, Gormley M, Paquet AC, Kjaer-Sorensen K, Kramer A, Rumer KK, Haimov-Kochman R, Yeh RF, Overgaard MT, Varki A, Oxvig C, Fisher SJ (2009) Severe preeclampsia-related changes in gene expression at the maternal–fetal interface include sialic acid-binding immunoglobulin-like lectin-6 and pappalysin-2. Endocrinology 150:452–462
Wishcamper CA, Coffin JD, Lurie DI (2001) Lack of the protein tyrosine phosphatase SHP-1 results in decreased numbers of glia within the motheaten (me/me) mouse brain. J Comp Neurol 441:118–133
Wood B (2002) Hominid revelations from Chad. Nature (Lond) 418:133–135
Wood B, Collard M (1999) The human genus. Science 284:65–71
Yamanaka M, Kato Y, Angata T, Narimatsu H (2009) Deletion polymorphism of SIGLEC14 and its functional implications. Glycobiology 19:841–846
Yousef GM, Ordon MH, Foussias G, Diamandis EP (2002) Genomic organization of the siglec gene locus on chromosome 19q13.4 and cloning of two new siglec pseudogenes. Gene (Amst) 286:259–270
Zhang JQ, Nicoll G, Jones C, Crocker PR (2000) Siglec-9, a novel sialic acid binding member of the immunoglobulin superfamily expressed broadly on human blood leukocytes. J Biol Chem 275:22121–22126
Author information
Authors and Affiliations
Corresponding author
Editor information
Editors and Affiliations
Rights and permissions
Copyright information
© 2012 Springer
About this chapter
Cite this chapter
Hayakawa, T., Varki, A. (2012). Human-Specific Changes in Sialic Acid Biology. In: Hirai, H., Imai, H., Go, Y. (eds) Post-Genome Biology of Primates. Primatology Monographs. Springer, Tokyo. https://doi.org/10.1007/978-4-431-54011-3_8
Download citation
DOI: https://doi.org/10.1007/978-4-431-54011-3_8
Published:
Publisher Name: Springer, Tokyo
Print ISBN: 978-4-431-54010-6
Online ISBN: 978-4-431-54011-3
eBook Packages: Biomedical and Life SciencesBiomedical and Life Sciences (R0)