Summary
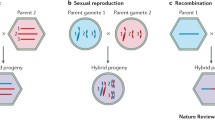
The Sindbis virus RNA genome is divided into two modules - one coding for the nonstructural protein genes and the other coding for the structural protein genes. In our studies of recombination, the two parental RNAs were defective in different modules. Analysis of the recombinant RNAs demonstrated that the parental RNAs each contributed its intact module and that the crossovers occurred within the defective modules. The recombinational events giving rise to infectious virion RNAs could create deletions, rearrangements or insertions as long as they occurred outside of the functional module. These crossovers produced RNA genomes that contained two functional subgenomic RNA promoters.
Chapter PDF
Similar content being viewed by others
Keywords
- Semliki Forest Virus
- Sindbis Virus
- Barley Stripe Mosaic Virus
- Eastern Equine Encephalitis Virus
- Brome Mosaic Virus
These keywords were added by machine and not by the authors. This process is experimental and the keywords may be updated as the learning algorithm improves.
References
Allison R, Thompson C, Ahlquist P (1990) Regeneration of a functional RNA virus genome by recombination between deletion mutants and requirement for cowpea chlorotic mottle virus 3a and coat genes for systemic infection. Proc Natl Acad Sci USA 87: 1820–1824
Bujarski JJ, Kaesberg P (1986) Genetic recombination between RNA components of a multipartite plant virus. Nature 321: 528–531
Edwards MC, Petty ITD, Jackson AO (1992) RNA recombination in the genome of barley stripe mosaic virus. Virology 189: 389–392
Gaedigk-Nitschko K, Schlesinger MJ (1991) Site-directed mutations in Sindbis virus E2 glycoprotein’s cytoplasmic domain and the 6K protein lead to similar defects in virus assembly and budding. Virology 183: 206–214
Geigenmüller-Gnirke U, Weiss B, Wright R, Schlesinger S (1991) Complementation between Sindbis viral RNAs produces infectious particles with a bipartite genome. Proc Natl Acad Sci USA 88: 3253–3257
Hahn CS, Lustig S, Strauss EG, Strauss JH (1988) Western equine encephalitis virus is a recombinant virus. Proc Natl Acad Sci USA 85: 5997–6001
Hirst GK (1962) Genetic recombination with Newcastle disease virus, polio- viruses, and influenza. Cold Spring Harbor Symp Quant Biol 27: 303–308
Jarvis TC, Kirkegaard K (1992) Polio virus RNA recombination: mechanistic studies in the absence of selection. EMBO J 11: 3135–3145
Khatchikian D, Orlich M, Rott R (1989) Increased viral pathogenicity after insertion of a 28S ribosomal RNA sequence into the haemagglutinin gene of an influenza virus. Nature 340: 156–157
King AMQ (1988) Genetic recombination in positive strand RNA viruses. In: Domingo E, Holland JJ, Ahlquist P (eds) RNA genetics, vol 2. CRC Press, Boca Raton, pp 150–185
King AMQ, McCahon D, Slade WR, Newman JWI (1982) Recombination in RNA. Cell 29: 921–928
Kirkegaard K, Baltimore D (1986) The mechanism of RNA recombination in poliovirus. Cell 47: 433–443
Lai MMC (1992) RNA recombination in animal and plant viruses. Microbiol Rev 56: 61–79
Lai MMC, Baric RS, Makino S, Keck J, Egbert J, Leibowitz JL, Stohlman SS (1985) Recombination between nonsegmented RNA genomes of murine corona- viruses. J Virol 56: 449–456
Liljeström P, Garoff H (1991) A new generation of animal cell expression vectors based on the Semliki Forest virus replicon. Bio/Technology 9: 1356–1361
Luytjes W, Bredenbeek PJ, Noten AF, Horzinek MC, Spaan WJ (1988) Sequence of mouse hepatitis virus A59 mRNA 2: indications for RNA-recombination between Coronavirus and influenza C virus. Virology 166: 415–422
Meyers G, Tautz N, Dubovi EJ, Thiel H-J (1991) Viral cytopathogenicity correlated with integration of ubiquitin-coding sequences. Virology 180: 602–616
Monroe SS, Schlesinger S (1983) RNAs from two independently isolated defective interfering particles of Sindbis virus contain a cellular tRNA sequence at their 5′ ends. Proc Natl Acad Sci USA 80: 3279–3283
Rice CM, Levis R, Strauss JH, Huang HV (1987) Production of infectious RNA transcripts from Sindbis virus cDNA clones: mapping of lethal mutations, rescue of a temperature-sensitive marker, and in vitro mutagenesis to generate defined mutants. J Virol 61: 3809–3819
Schlesinger S, Schlesinger M (1990) Replication of togaviridae and flaviviridae. In: Fields BN, Knipe DM (eds) Virology. Raven Press, New York, pp 697–711
Strauss EG, Strauss JH (1986) Structure and replication of the alphavirus genome. In: Schlesinger S, Schlesinger MJ (eds) The togaviridae and flaviviridae. Plenum Press, New York, pp 35–82
Weiss BG, Schlesinger S (1991) Recombination between Sindbis virus RNAs. J Virol 65: 4017–4025
Xiong C, Levis R, Shen P, Schlesinger S, Rice CM, Huang HV (1989) Sindbis virus: an efficient, broad host range vector for gene expression in animal cells. Science 243: 1188–1191
Author information
Authors and Affiliations
Editor information
Editors and Affiliations
Rights and permissions
Copyright information
© 1994 Springer-Verlag
About this paper
Cite this paper
Schlesinger, S., Weiss, B.G. (1994). Recombination between Sindbis virus RNAs. In: Brinton, M.A., Calisher, C.H., Rueckert, R. (eds) Positive-Strand RNA Viruses. Archives of Virology Supplementum, vol 9. Springer, Vienna. https://doi.org/10.1007/978-3-7091-9326-6_21
Download citation
DOI: https://doi.org/10.1007/978-3-7091-9326-6_21
Publisher Name: Springer, Vienna
Print ISBN: 978-3-211-82522-8
Online ISBN: 978-3-7091-9326-6
eBook Packages: Springer Book Archive