Abstract
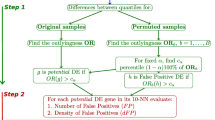
Cancer is a disease driven by pathway activity, while useful biomarkers to predict outcome (prognostic markers) or determine treatment (treatment markers) rely on individual genes, proteins, or metabolites. We provide a novel approach that isolates pathways of interest by integrating outlier analysis and gene set analysis and couple it to the top-scoring pair algorithm to identify robust biomarkers. We demonstrate this methodology on pediatric acute myeloid leukemia (AML) data. We develop a biomarker in primary AML tumors, demonstrate robustness with an independent primary tumor data set, and show that the identified biomarkers also function well in relapsed AML tumors.
Chapter PDF
Similar content being viewed by others
Keywords
- Acute Myeloid Leukemia
- Outlier Analysis
- Diagnostic Sample
- Pediatric Acute Myeloid Leukemia
- Relapse Sample
These keywords were added by machine and not by the authors. This process is experimental and the keywords may be updated as the learning algorithm improves.
References
Geman, D., d’Avignon, C., Naiman, D.Q., Winslow, R.L.: Classifying gene expression profiles from pairwise mrna comparison. Statistical Applications in Genetics and Molecular Biology 3(1), 19 (2004)
Gentleman, R.C., Carey, V.J., Bates, D.M., Bolstad, B., Dettling, M., Dudoit, S., Ellis, B., Gautier, L., Ge, Y., Gentry, J., Hornik, K., Hothorn, T., Huber, W., Iacus, S., Irizarry, R., Leisch, F., Li, C., Maechler, M., Rossini, A.J., Sawitzki, G., Smith, C., Smyth, G., Tierney, L., Yang, J.Y., Zhang, J.: Bioconductor: open software development for computational biology and bioinformatics. Genome Biol 5(10), R80 (2004)
Ghosh, D.: Discrete nonparametric algorithms for outlier detection with genomic data. Journal of Biopharmaceutical Statistics 20(2), 193–208 (2010)
Hanahan, D., Weinberg, R.A.: The hallmarks of cancer. Cell 100(1), 57–70 (2000)
Hanahan, D., Weinberg, R.A.: Hallmarks of cancer: the next generation. Cell 144(5), 646–674 (2011)
Kanehisa, M., Goto, S., Kawashima, S., Nakaya, A.: The KEGG databases at genomenet. Nucleic Acids Res. 30(1), 42–46 (2002)
Knox, S.S., Ochs, M.F.: Implications of systemic dysfunction for the etiology of malignancy. Gene. Regul. Syst. Bio. 7, 11–22 (2013)
Liberzon, A., Subramanian, A., Pinchback, R., Thorvaldsdottir, H., Tamayo, P., Mesirov, J.P.: Molecular signatures database (MSigDB) 3.0. Bioinformatics 27(12), 1739–1740 (2011)
MacDonald, J.W., Ghosh, D.: COPA–cancer outlier profile analysis. Bioinformatics 22(23), 2950–2951 (2006)
Michaud, J., Simpson, K.M., Escher, R., Buchet-Poyau, K., Beissbarth, T., Carmichael, C., Ritchie, M.E., Schütz, F., Cannon, P., Liu, M., Shen, X., Ito, Y., Raskind, W.H., Horwitz, M.S., Osato, M., Turner, D.R., Speed, T.P., Kavallaris, M., Smyth, G.K., Scott, H.S.: Integrative analysis of RUNX1 downstream pathways and target genes. BMC Genomics 9, 363 (2008)
Parsons, D.W., Jones, S., Zhang, X., Lin, J.C., Leary, R.J., Angenendt, P., Mankoo, P., Carter, H., Siu, I.M., Gallia, G.L., Olivi, A., McLendon, R., Rasheed, B.A., Keir, S., Nikolskaya, T., Nikolsky, Y., Busam, D.A., Tekleab, H., Diaz Jr., L.A., Hartigan, J., Smith, D.R., Strausberg, R.L., Marie, S.K., Shinjo, S.M., Yan, H., Riggins, G.J., Bigner, D.D., Karchin, R., Papadopoulos, N., Parmigiani, G., Vogelstein, B., Velculescu, V.E., Kinzler, K.W.: An integrated genomic analysis of human glioblastoma multiforme. Science 321(5897), 1807–1812 (2008)
Price, N.D., Trent, J., El-Naggar, A.K., Cogdell, D., Taylor, E., Hunt, K.K., Pollock, R.E., Hood, L., Shmulevich, I., Zhang, W.: Highly accurate two-gene classifier for differentiating gastrointestinal stromal tumors and leiomyosarcomas. Proc. Natl. Acad. Sci. U S A 104(9), 3414–3419 (2007)
Smyth, G.K.: Limma: linear models for microarray data, pp. 397–420. Springer, New York (2005)
Tan, A.C., Naiman, D.Q., Xu, L., Winslow, R.L., Geman, D.: Simple decision rules for classifying human cancers from gene expression profiles. Bioinformatics 21(20), 3896–3904 (2005)
Author information
Authors and Affiliations
Editor information
Editors and Affiliations
Rights and permissions
Copyright information
© 2013 Springer-Verlag Berlin Heidelberg
About this paper
Cite this paper
Ochs, M.F., Farrar, J.E., Considine, M., Wei, Y., Meschinchi, S., Arceci, R.J. (2013). Outlier Gene Set Analysis Combined with Top Scoring Pair Provides Robust Biomarkers of Pathway Activity. In: Ngom, A., Formenti, E., Hao, JK., Zhao, XM., van Laarhoven, T. (eds) Pattern Recognition in Bioinformatics. PRIB 2013. Lecture Notes in Computer Science(), vol 7986. Springer, Berlin, Heidelberg. https://doi.org/10.1007/978-3-642-39159-0_5
Download citation
DOI: https://doi.org/10.1007/978-3-642-39159-0_5
Publisher Name: Springer, Berlin, Heidelberg
Print ISBN: 978-3-642-39158-3
Online ISBN: 978-3-642-39159-0
eBook Packages: Computer ScienceComputer Science (R0)