Abstract
Endogenous retroviruses have traditionally been defined as descendants of extinct retroviruses that infected and integrated into the chromosomes of host germ-line cells and were thereafter transmitted vertically as part of host genomes. Most retain at least the vestiges of genes once required for infectious horizontal transfer, namely envelope genes. In contrast, the long evolutionary histories of retrotransposons are presumed not to have included infectious ancestors. With the characterization of the Gypsy retrotransposon in Drosophila melanogaster as an infectious, endogenous retrovirus, these distinctions have blurred. A number of plant LTR retroelements possess coding regions whose conceptual translations produce hypothetical proteins with predicted structural elements found in viral envelope proteins, and the term endogenous retrovirus began to be applied to these elements. The question of whether any of the many plant retroelement genes now annotated as “env-like” generate proteins that have or had envelope functions remains unanswered. This review reevaluates the available data.
You have full access to this open access chapter, Download chapter PDF
Similar content being viewed by others
Keywords
6.1 Beyond gag and pol: Plant Retroelements with Extra ORFs
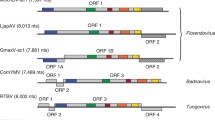
While plant LTR retrotransposons are generally easily identified by conserved domains in the POL polyprotein [retropepsin (PROT), integrase (INT), reverse transcriptase (RT), and RNase H (RH)], and to a lesser extent by zinc knuckle RNA-binding motifs in GAG, there are a significant number of families among both Ty3/Gypsy and Ty1/Copia superfamilies that possess additional or extra open reading frames (eORFs) (Fig. 6.1). The conceptual translations of most of these eORFs produce novel proteins with no definitive homology to proteins with known functions (Peterson-Burch et al. 2000; Wicker and Keller 2007; Grandbastien 2008; Steinbauerová et al. 2012), nor have protein products from these eORFs been isolated, let alone functionally assayed. In most cases, these regions are found between pol and the 3′ LTR (3′ eORFs) (Fig. 6.1), but there are several exceptions (Steinbauerová et al. 2012). Members of the Ogre lineage, best characterized in legumes, possess conserved, intact 5′ eORFs between the 5′ LTR and gag (Fig. 6.1) (Neumann et al. 2003; Macas and Neumann 2007; Steinbauerová et al. 2012).
Structure of LTR retroelements with extra ORFs. LTRs, black triangles; extra 5′ ORF, blue arrow; gag, brown arrow; pol, green arrow; 3′ extra ORF, red arrow; black dots, noncoding regions. gag and pol may be fused and translated in a single reading frame, separated by a stop codon or in different reading frames. Distances between elements are variable. Not to scale
There are a few instances of small numbers of elements containing fragments of recognizable host genes, the probable result of transcriptional readthrough or recombinational capture (Jin and Bennetzen 1994; Du et al. 2006; SanMiguel and Vitte 2009; Steinbauerová et al. 2012). It is doubtful these host genes played any functional role, and these elements will not be addressed here. Interestingly, Steinbauerová et al. (2012) reported partial sequence similarities in eORFs to the plant mobile domain, a member of a group of conserved zinc finger motifs found in a large superfamily of eukaryotic transcription factors and shown to be associated with MULE transposases (Babu et al. 2006). These similarities were found within 5′ eORFs or 3′ eORFs in a single clade of Ty3/Gypsy elements that included the Ogre family. Finally, the DIRS-1 retrotransposon family is characterized by a domain encoding a tyrosine recombinase at the 3′ end of pol (Poulter and Goodwin 2005; Wicker and Keller 2007), but no representatives have been found in plants (Piedöel et al. 2011).
The partial conservation of the conceptual translations of some 3′ eORFs in several retroelement families in species as distantly related as Arabidopsis, tomato, soybean, maize, and barley strongly suggests that these proteins play or have played an important role in the proliferation of these elements. What that role or roles may be is open to speculation, but for reasons that will be discussed below, many of these eORFs were described as “envelope-like” based on varying degrees of predicted secondary structure similarity of their conceptual translation products to viral envelope proteins (Laten et al. 1998; Peterson-Burch et al. 2000; Vicient et al. 2001; Wright and Voytas 2002; Boeke et al. 2005b; Holligan et al. 2006; Hafez et al. 2009; Laten and Bousios 2012). By extension, it has been suggested that these retrotransposon families are analogous to animal endogenous retroviruses (Kumar 1998; Laten et al. 1998; Peterson-Burch et al. 2000; Wright and Voytas 2002), the integrated vestiges of ancient infectious retroviruses.
6.2 Viral Envelope Proteins
Viral envelope proteins are a diverse family of glycoproteins that sponsor attachment, entry into, and exit from infected cells by “enveloped” viruses like Influenza A, Hepatitis C, SARS Coronavirus, and HIV (Harrison 2008; Cosset and Lavillette 2011). These processes include peptide cleavage, receptor binding, intracellular targeting and transport, disulfide bond formation, glycosylation, membrane fusion, and oligomerization (Cosset and Lavillette 2011). In the case of many, including those of retroviruses, structural features of the envelope protein may include a signal peptide, a proline-rich region, transmembrane domains, a coiled coil, a fusion peptide, and a conserved cleavage site (Wu et al. 1998; Harrison 2008; Cosset and Lavillette 2011) (Fig. 6.2). Many viral envelope proteins, including those of retroviruses, are translated as precursors that are cleaved into a surface glycoprotein and a transmembrane glycoprotein (Hunter 1997) (see Fig. 6.2).
6.2.1 Envelope Protein Variation
While structural and functional elements are shared by diverse groups of viral envelope proteins, amino acid sequence variation is high, and it remains unclear if the three major classes of envelope proteins—based on their fusion peptides—are related by descent from a single ancestral gene (Kadlec et al. 2008; Cosset and Lavillette 2011). For mammalian retroviruses and endogenous retroviruses, even in cases where clear evolutionary relationships are inferred from phylogenetic trees based on RT alignments, the corresponding envelope sequences may have diverged to the extent that homology cannot be deduced from global sequence-based analyses (Benit et al. 2001). However, by restricting multi-sequence alignments to transmembrane subunits, homology has been inferred across retroviral genomes and those of other enveloped viruses, such as Ebola and Marburg, with Class I fusion proteins (Benit et al. 2001). Although not addressed by Benit et al. (2001), the observed localized sequence similarities could have been the result of convergent evolution or localized domain capture.
Kim et al. (2004) suggested that the homology between distantly related retroviruses is the result of envelope capture, and this hypothesis is supported by the phylogenetic analysis of Benit et al. (2001). The origins of these envelope genes are unknown. Furthermore, envelope capture is not unique to vertebrate viruses (Pearson and Rohrmann 2002) (see below).
In mammals, viral envelope variation in the surface protein subunit is likely driven to a large degree by positive selection in response to host adaptive immune systems (Caffrey 2011). While innate immune responses in vertebrates, invertebrates, and plants have been shown to contribute to the evolution of virulence/effector proteins in pathogens that attenuate these responses (Finlay and McFadden 2006; Nishimura and Dangl 2010), there is no evidence that antigenic variation is employed as a mechanism to escape innate immunity (Finlay and McFadden 2006). Nor is there any evidence that envelope variants are responsible for suppression or evasion of silencing of viral gene expression by host siRNAs in plants or animals (Li and Ding 2006; Obbard et al. 2009).
6.3 Endogenous Retroviruses
Endogenous retroviruses (ERV) are the integrated remains of extinct retroviruses that infected and reinfected host germ-line cells, inserting into germ-line chromosomes and consequently vertically inherited by generations of host descendants (Bannert and Kurth 2006; Jern and Coffin 2008; Ribet et al. 2008; Feschotte and Gilbert 2012).
6.3.1 Human and Other Vertebrate Endogenous Retroviruses
With the possible exception of the highest copy-number families (Belshaw et al. 2005), few endogenous human retroviruses appear to be capable of autonomous retrotransposition in germ-line cells (Belshaw et al. 2004), most likely because of debilitating mutations and/or epigenetic silencing (Belshaw et al. 2005; Maksakova et al. 2008). However, some murine ERVs are far more active (Maksakova et al. 2006). In most ERV families, the envelope gene sequences are riddled with nonsense mutations and deletions. It has been suggested that most, but not all, vertebrate multi-copy ERV families arose by short bursts of multiple germ-line infections, not by retrotransposition (Belshaw et al. 2004; Bannert and Kurth 2006; Jern and Coffin 2008). While there is no evidence for recent retrotransposition of human ERVs, mobilization of ERVs in other mammals has been reported (Maksakova et al. 2006, 2008; Ribet et al. 2007; Stocking and Kozak 2008; Zhang et al. 2008; Wang et al. 2010), and the expression of ERV mRNA and production of proteins in somatic tissue has been associated with some cancers (Moyes et al. 2007; Howard et al. 2008; Maksakova et al. 2008).
6.3.2 Invertebrate Endogenous Retroviruses
Env-like genes downstream of pol have been reported for several invertebrate LTR-retroelements. Most notably, Gypsy from D. melanogaster has long been recognized as an endogenous retrovirus (Kim et al. 1994; Song et al. 1994) with strong evidence that it retains infectivity (Kim et al. 1994; Song et al. 1994; Teysset et al. 1998; Pelisson et al. 2002; Misseri et al. 2004). While transfer of Gypsy elements from somatic to germ-line tissue does not require a functional env gene (Chalvet et al. 1999), the Gypsy envelope glycoprotein has been shown to sponsor cell–cell fusion in cell culture assays (Misseri et al. 2004). Other invertebrate retroelements that contain envelope-like coding regions include several additional Drosophilid elements (Mejlumian et al. 2002; Llorens et al. 2008, 2011), TED, a lepidopteran element from Trichoplusia ni (Friesen and Nissen 1990; Ozers and Friesen 1996), yoyo from the Mediterranean fruit fly, Ceratitis capata (Zhou and Haymer 1998), Tas from the parasitic nematode Ascaris lumbricoides (Felder et al. 1994), Cer7 (Bowen and McDonald 1999) from C. elegans, and two elements, Juno and Vesta, from bdelloid rotifers (Gladyshev et al. 2007).
The env-like regions of the insect elements have been shown to be homologous (Terzian et al. 2001). Many of the hypothetical ENV-like proteins contain multiple structural features common to viral envelope proteins. Based on sequence similarities, Eickbush and Malik (2002) suggested that the env-like genes in Tas and Cer7 were derived from a Phlebovirus and a Herpesvirus, respectively. With the exception of Gypsy, invertebrate retroelements have not been demonstrated to be infectious. Gypsy and related arthropod elements have been designated as Errantiviruses (Boeke et al. 2005a).
Several phylogenetic and functional analyses strongly suggest that the genes encoding the Errantivirus envelope-like proteins are derived from Baculoviral env genes (Malik et al. 2000; Rohrmann and Karplus 2001; Pearson and Rohrmann 2002, 2004, 2006; Misseri et al. 2003; Kim et al. 2004). However, any homology to vertebrate retroviral envelope proteins is only weakly supported at best (Lerat and Capy 1999; Malik et al. 2000), and the very small number of short blocks of amino acid similarity between conserved Errantivirus envelope proteins and those of vertebrate retroviruses could be fortuitous, or the result of convergent evolution or recombinational domain capture.
6.3.3 Are There Plant Endogenous Retroviruses?
Animal endogenous retroviruses have been defined as vertically transmitted, retroviral-related DNAs distinguished from LTR retrotransposons by the presence of at least vestiges of an envelope-coding region downstream of pol and/or a close phylogenetic relationship to extant retroviruses (Boeke and Stoye 1997; Bannert and Kurth 2006; Jern and Coffin 2008; Feschotte and Gilbert 2012). In the case of plants, infectious retroviruses have not been reported. However, integrated, vertically transmitted copies of plant pararetroviral genomes are widespread in both dicots and monocots (Staginnus and Richert-Poggeler 2006; Hohn et al. 2008). Plant pararetroviruses, like the Caulimoviruses, are DNA viruses characterized by genomes encoding GAG, PROT, RT, and RH, as well as additional essential proteins (Lazarowitz 2007). Unlike retroviruses, pararetroviruses are not enveloped, and their infectious cycles do not normally include integration into the host genome (Lazarowitz 2007). Integration appears to be extremely rare, and integrated viral sequences are generally incomplete, rearranged and mutated, and not known to be infectious or capable of autonomous retrotransposition (Staginnus and Richert-Poggeler 2006; Hohn et al. 2008).
The first suggestions that plant genomes might contain endogenous retroviruses were made based on the presence of predicted ENV-like structural features in the conceptual translations of LTR elements with 3′ eORFs of several hundreds to over 2,000 bp (Laten et al. 1998; Wright and Voytas 1998). Four families of Athila elements, members of the Ty3/Gypsy superfamily from A. thaliana, were initially shown to contain extended ORFs downstream of int with conceptual translation products containing one or more predicted transmembrane regions (Wright and Voytas 1998). These sequences were not considered to be homologous to retroviral env genes, but the suggestion was made that the encoded proteins might once have promoted membrane fusion (Wright and Voytas 1998).
Predicted structural similarities between viral envelope proteins and the conceptual translation of a 3′ eORF of an unrelated element, SIRE1 from Glycine max, were far more extensive (Laten et al. 1998). The suggestion that SIRE1, a member of the Ty1/Copia superfamily, encoded an envelope-like protein was derived from several features of the conceptual translation of the long, uninterrupted 3′ eORF in the same reading frame as pol but separated from pol by a single stop codon. The conceptual translation of this ORF produced a 70 kDa, 650-amino acid polypeptide (Laten et al. 1998). This hypothetical protein was predicted to contain transmembrane domains at positions corresponding to the signal and fusion domains of viral envelope proteins and a strongly predicted coiled coil in a region corresponding to those containing coiled coils in several viral envelope proteins, including that of HIV (Laten et al. 1998) (Fig. 6.2). While the conceptual translation contained only two N-glycosylation motifs, there were several serines and threonines in contexts known to promote O-glycosylation, a characteristic of many viral envelope proteins (Pinter and Honnen 1988; Wilson et al. 1991). In addition, there was an extended proline-rich region from amino acid 60 to 128. The overall amino acid composition of this region was remarkably similar to those found in the neutralization domains of some mammalian retroviruses (Laten et al. 1998).
Retroviral envelope proteins are known to be expressed from spliced transcripts (Rabson and Graves 1997). However, there are no recognizable splice acceptor sites in SIRE1 or in related elements that would fuse this ORF with an upstream start codon (Peterson-Burch and Voytas 2002). Nor are there AUG codons downstream of the pol stop codon that might support translational initiation at an internal ribosomal entry site (Peterson-Burch and Voytas 2002). However, Havecker and Voytas (2003) showed that the SIRE1 pol stop codon was embedded in a hexanucleotide motif that had previously been shown to sponsor developmentally regulated stop codon suppression in tobacco mosaic virus and in yeast. They demonstrated that the SIRE1 sequence supported low levels of stop codon suppression (5%) in in vivo readthrough assays and that suppression was lost with single base-pair changes in the sequence (Havecker and Voytas 2003).
Once the potential characteristics of these unusual elements were recognized, analyses of previously reported plant retrotransposons with long uncharacterized regions between pol and the 3′ LTR revealed that conceptual translation of these interrupted 3′ eORFs could generate hypothetical proteins with highly significant sequence similarity to those described above (Laten 1999; Peterson-Burch et al. 2000) (see Table 6.1). Three of these hypothetical proteins were aligned to highlight their similarities (Fig. 6.3). The extent and degree of sequence identity was variable but in the case of SIRE1 and Endovir1 encompassed most of the sequence. The densities of sequence matches were far greater in the second half of the alignment. The distances between the pol stop codon and the beginning of the env-like coding region were also highly variable, ranging from 0 to over 1,000 bp (Peterson-Burch and Voytas 2002; Laten et al. 2003; Havecker et al. 2005; Weber et al. 2010).
Alignment of ENV-like regions from ToRTL1 from S. lycopersicum, Endovir1-1 from A. thaliana, and SIRE1 from G. max. The env-like ORFs are represented by white bars and are drawn to scale. Black lines depict noncoding sequences between pol and the start of the env-like ORF. Regions of amino acid similarity between elements are connected by shading. Percentages on the left represent the total amino acid similarity over the shaded regions. The numbers of amino acids in the env-like ORFs are given for each element. Predicted features are denoted as follows: α-helices, dark gray boxes; β-sheets, arrows; transmembrane domains, slanted line boxes. Adapted from Peterson-Burch and Voytas (2002) with permission
The phylogenetic relationships among groups of retroelements with and without eORFs are illustrated in Fig. 6.4. A fusion of the network analyses of Llorens et al. (2009) and the more classical approach illustrated in Eickbush and Jamburuthugoda (2008), this consensus tree illustrates the widespread acquisition of primarily 3′ eORFs with both known, as in the case of vertebrate retroviruses and Gypsy, and unknown function.
(a) Simplified, unrooted phylogeny of LTR-related retroelements. Modeled with modification after Eickbush and Jamburuthugoda (2008) and Llorens et al. (2009). Branch lengths do not represent distances. (b) Presence of eORFs in one or more members within terminal clades representing groups of related subfamilies indicated with Y. Absence of eORF in all subfamilies within a terminal clade indicated with N. Metaviridae family defined by Boeke et al. (2005a). Pseudoviridae family defined by Boeke et al. (2005b). Data sources for B: Llorens et al. 2011; Steinbauerová et al. 2012; King et al. 2012 (http://ictvonline.org/index.asp)
6.3.3.1 Ty1/Copia Sireviruses
The SIRE1 element family in soybean, with as many as 1,350 copies per genome (Laten and Morris 1993; Du et al. 2010b; Bousios et al. 2012b), is highly conserved and recently amplified (Laten et al. 2003; Du et al. 2010b; Bousios et al. 2012b). Nearly all copies have inserted into their present genomic positions in the last 750,000 years, with as many as 10% having done so in the last 30,000 (Du et al. 2010b; Bousios et al. 2012b). SIRE1 has been designated as the Type Species for the Genus Sirevirus (Boeke et al. 2005b), and based on reverse transcriptase sequences constitutes a monophyletic group within the Ty1/Copia superfamily (Boeke et al. 2005b; Du et al. 2010b; Bousios et al. 2012a). This group has been alternatively designated as the Maximus lineage (Du et al. 2010b) or the Sirevirus lineage (Bousios et al. 2012a). Not all members of the lineage contain 3′ eORFs that encode hypothetical proteins with ENV-like features (Havecker et al. 2005; Pearce 2007; Bousios et al. 2010, 2012a, b), but those that do have been found in the genomes of most eudicots and monocots for which extensive sequence data are available (see Table 6.1). Many of the hypothetical proteins are truncated or heavily mutated and have not been annotated. The initial recognition and discovery of some of these 3′ eORFs required tBLASTn searches of nucleotide databases using previously reported ENV-like proteins as queries (Laten 1999; Havecker et al. 2005; Wicker and Keller 2007; Du et al. 2010b; Laten and Bousios 2012).
Recognizable conservation of the ENV-like peptide sequences extends to a broad range of eudicot taxa and includes members in the order Fabales, Vitales, Brassicales, Solanales, Lamiales, and Caryophyllales. Most of the extended sequence identities and similarities shared by these hypothetical proteins would correspond to the carboxyl half of a retroviral protein encompassing the transmembrane protein and part of the surface protein (see Fig. 6.2). However, not all of these hypothetical proteins contain predicted transmembrane domains (Havecker et al. 2005) (Fig. 6.5), and, not unexpectedly, multi-sequence alignments generated few positions with consensus residues (Havecker et al. 2005). Weaker sequence similarity corresponding to the first 300 amino acids of the SIRE1 ENV-like hypothetical protein has only been detected in short regions of the related elements in L. japonicus (Laten, unpublished). Additional members of the same lineage, based on their RT sequences, possess several hundred bp between the pol stop codon and the 3′LTR, including PREM-2, Opie-2, and most members of the Ji lineage from maize, and Osr7 and Osr8 from rice. These elements have no discernible 3′ eORFs, although the maize Jienv clade does (Bousios et al. 2012a).
Predicted structural elements found in translated 3′ ORFs of selected members of the Sirevirus family. Adapted from Havecker et al. (2005) with permission
Even among the elements for which env-like ORFs have been deduced, few Sireviruses with intact env-like regions with greater than 500 contiguous codons have been found. The recognition of others are often derived from consensus sequences generated from multi-sequence alignments (Wicker and Keller 2007; Laten et al. 2009). Among those that possess long intact 3′ eORFs, the G. max, L. japonicus, B. vulgaris, and M. guttatus Sireviruses encode hypothetical ENV proteins of 648–680, 630–949, 606, and 780 amino acids, respectively, for SIRE1 (Laten et al. 2003), Lotus2 (Holligan et al. 2006), Cotzilla1 (Weber et al. 2010), and MguSIRV (Laten and Bousios 2012).
Neighbor joining trees of Sirevirus RT domains showed that those elements containing intact or vestiges of “ENV-like” domains appear to be monophyletic (Bousios et al. 2010, 2012a; Du et al. 2010b). Members of the Maximus lineage (Wicker and Keller 2007) all fall within the Sirevirus clade based on their RT domains (Fig. 6.6) (Bousios et al. 2010; Du et al. 2010b) and most are characterized by extended GAG regions with multiple RNA binding motifs and predicted coiled coils (Peterson-Burch and Voytas 2002; Havecker et al. 2005). Bousios et al. (2010) have also described a number of highly conserved features in Sirevirus noncoding regions in the LTR and immediately upstream of the 3′ LTR.
Neighbor joining phylogenetic tree based on shared RT/RH domains highlighting the Sirevirus clade. From Bousios et al. (2010)
The Sirevirus group in L. japonicus is the predominant Ty1/Copia lineage in L. japonicus, constituting 40% of these retroelements (Holligan et al. 2006). This group is also among the most recently amplified in the L. japonicus genome, with many members possessing identical LTR sequences (Holligan et al. 2006). As in the case of SIRE1, most of the full-length elements in this lineage contain intact 3′ eORFs ranging in length from 630 to 949 codons. The conceptual translation products in two of three sub-lineages contained predicted transmembrane domains and the product of one sub-lineage also contained a predicted coiled coil (Holligan et al. 2006). However, Holligan et al. (2006) reported that significant similarities among the ENV-like sequences were restricted to the individual sub-lineages.
SIRE is also the predominant retroelement in the Ty1/Copia lineage in G. max (Du et al. 2010b), and the Maximus lineage is the predominant retroelement group in banana, constituting 13% of that genome (Hribova et al. 2010). The Osr8 lineage in the Sirevirus clade (Fig. 6.6) is also the most abundant Ty1/Copia lineage in the rice genome (McCarthy et al. 2002).
In the maize genome, retroelement families identified as members of the Sirevirus lineage with ENV-like domains, Hopie, Giepum, and Jienv, and those without, Opie and Ji, are represented by >10,600 intact and approximately 28,000 degenerate copies (Bousios et al. 2012a). This constitutes as much as 90% of the total population of Ty1/Copia elements in maize. Many of these insertions occurred within the last 600,000 years (Bousios et al. 2012a).
Cotzilla1 from B. vulgaris is another recently reported member of the Sirevirus genus (Weber et al. 2010). Conceptual translation of its env-like gene generates a proline-rich region and a predicted coiled coil near the carboxyl terminal but no predicted transmembrane domains (Weber et al. 2010). The 606-codon env-like ORF begins 561 bp downstream from the end of pol. With an estimated copy number of 2,100 and members as young or younger than 290,000 years, Cotzilla may be the youngest and most abundant retroelement family in the sugar beet genome (Weber et al. 2010).
The lineages containing G. max and L. japonicus are estimated to have separated from each other over 50 million years ago (Lavin et al. 2005). In addition to the genus Lotus, the latter lineage contains the genera Medicago, Pisum, and Trifolium. While the species in these genera contain Sirevirus-like sequences with at least fragments of homologous env-like ORFs, fully intact env-like ORFs have not been reported. The relative youth of the apparently functional copies of the Sireviruses in G. max and L. japonicus suggests that significant amplification of one or a few ancestral copies with preexisting intact env-like ORFs occurred over the last few hundreds of thousands of years, with integration of some copies of diverged sub-lineages within the last tens of thousands years (Laten et al. 2003; Holligan et al. 2006; Du et al. 2010b). The presence of intact or nearly intact retroelement 3′ eORFs that have retained and/or acquired shared predicted structural elements over such a broad range of taxa argues strongly for function. However, expression of these elements has not been unequivocally demonstrated.
In the case of SIRE1, transcripts were not detected in northern blots, but gag, rt and env transcripts were detected by RT-PCR of leaf and/or root tissue (Lin 2001). However, amplification of RNAs derived from high copy-number elements does not signify functional expression because of the strong possibility of cryptic transcriptional initiation or readthrough sponsored by adjacent promoters. The 30 EST sequences containing SIRE1 fragments in the Genbank database as of May 2011 are equally distributed among sense and antisense transcripts (Gaston 2011).
The SIRE1 env-like ORF has been expressed from fusion constructs in S. cerevisiae (Gouvas and Laten, unpublished) and in E. coli (Gaston 2011). In the case of the former, yeast two-hybrid screens suggested that the protein self-associates and forms protein–protein interactions with at least two other soybean proteins with transmembrane domains (Gouvas and Laten, unpublished). In preliminary experiments, polyclonal antibodies raised against a sub-region expressed in E. coli bound to a 65-kDa protein isolated from soybean callus tissue (Gaston 2011). The protein has not been identified, but is only slightly smaller than the 70 kDa predicted for the SIRE1-4 ENV.
6.3.3.2 Plant Ty3/Gypsy “Endogenous Retroviruses”
The number of plant Ty3/Gypsy elements characterized as encoding ENV-like proteins is presently fewer than that in the Sirevirus lineage but just as widely distributed among taxa (Grandbastien 2008). As in the case of the Sireviruses, there is considerable variation in the amino acid sequences of the conceptually translated ORFs and in the possession of ENV-like secondary structures in elements from Arabidopsis (Wright and Voytas 1998, 2002), soybean (Wright and Voytas 2002; Du et al. 2010b), pea (Neumann et al. 2005), and barley (Vicient et al. 2001). These include transmembrane domains, coiled coils, cleavage sites, and N-glycosylation motifs. Many other elements within the same lineages possess vestiges of these regions that can be shown to be related through tBLASTn searches (e.g., Neumann et al. 2005). With the exception of one family (see below), all fall within the Athila clade based on their RT sequences.
The Athila family itself was the first among plant elements in the Ty3/Gypsy superfamily to be labeled as possible endogenous retroviruses based on the presence of 3′ eORFs whose conceptual translation produced hypothetical proteins with strongly predicted, transmembrane domains (Wright and Voytas 1998, 2002). Although highly degenerate, consensus elements were constructed for seven subfamilies, and all contained ENV-like hypothetical proteins with at least one predicted transmembrane domain (Wright and Voytas 2002). In addition, splice acceptor sites were predicted near the beginning of the 3′ eORF (Wright and Voytas 1998, 2002). The Athila4 consensus generated a 619-amino acid ENV-like hypothetical protein (Wright and Voytas 2002).
With the recognition that 3′ eORFs in Ty3/Gypsy elements might encode ENV-like proteins based on shared predicted secondary structural elements, related elements were sought and found in a broad range of taxa beginning with two related element families: Cyclops-2 in P. sativum (Chavanne et al. 1998; Peterson-Burch et al. 2000) and the Calypso family in G. max (Peterson-Burch et al. 2000; Wright and Voytas 2002) (see Table 6.1). The env-like ORF in the former was 423 codons and 420 in the latter. As in the case of Athila, Calypso had a strongly predicted splice acceptor site near the 5′ end of the env-like ORF (Wright and Voytas 2002). Analyses of the transmembrane domains suggested targeting to the plasma membrane in the case of Calypso2 and the endoplasmic reticulum in the case of Athila4 (Wright and Voytas 2002).
While individual members of the Athila and Calypso families are degenerate and appear to be nonfunctional, a related family in barley, Bagy-2, contains copies with intact ORFs for gag and pol, and an intact env-like ORF whose conceptual translation produces a 47-kDa protein (Fig. 6.7) (Vicient et al. 2001). Furthermore, RT-PCR amplification from several tissues with 3′ eORF-specific primers suggested that Bagy-2 is transcribed and that transcripts are spliced (Vicient et al. 2001). In addition, insertional polymorphisms among a number of related barley cultivars suggested that Bagy-2 copies have recently transposed (Vicient et al. 2001). A consensus sequence for a closely related element with an ENV-like hypothetical protein in rice, Rigy-2, was generated from an alignment of four copies interrupted by other nested elements (Fig. 6.6). The 3′ eORFs in the Rigy-2 consensus sequence contained both nonsense and frameshift mutations (Vicient et al. 2001). Related elements have also been reported in cultivated allotetraploid cotton and their diploid progenitors, and the hypothetical ENV-like proteins are strongly predicted to possess transmembrane domains (Hafez et al. 2009).
Features of the Bagy-2 and Rigy-2 retrovirus-like retrotransposons and their predicted ENV-like attributes. Putative N-glycosylation sites (↑), proteinase cleavage site (¥), leucine zipper (LZ), and transmembrane domains (TM). From Vicient et al. (2001) with permission
TBLASTn searches using the Bagy-2 ENV hypothetical protein retrieved statistically significant hits (e < 10−8) to sequences in several legume species (M. truncatula, L. japonicus, G. max, V. radiata and V. unguiculata, T. pratense, A. duranensis, C. cajan, P. vulgaris, and T. labialis), and in carrot (D. carota), monkey flower (M. guttatus and M. lewisii), tobacco (N. tabacum), and ginseng (P. ginseng) (Laten, unpublished).
The PIGY family from P. sativum also contains members with 3′ eORFs whose conceptual translations produce hypothetical proteins with predicted transmembrane domains. These showed significant amino acid similarity to the Athila ENV-like hypothetical proteins (Neumann et al. 2005). A related but highly disrupted family, MEGY, was also found in M. truncatula (Neumann et al. 2005).
Another related element family, FIDEL, has recently been characterized from peanut (Nielen et al. 2010). The 3′ LTR of FIDEL is separated from the end of pol by 2.1 kb, but no members of this family contained an extended ORF in this region (Nielen et al. 2010). However, conceptual translations of this region in a FIDEL consensus sequence generated multiple, strongly predicted transmembrane domains (Laten, unpublished).
As in the case of the Sireviruses, most of the 3′ eORFs from these elements—all members of the Athila clade (Llorens et al. 2011)—are interrupted by multiple stop codons and/or frameshifts, and recognition of amino acid sequence conservation across families is often difficult. Nonetheless, these regions appear to have been under some degree of negative selection during their evolutionary history (Vicient et al. 2001; Wright and Voytas 2002; Neumann et al. 2005).
Families in the Tat clade, which include Grande1, Tat4, RIRE2, Ogre, RetroSort, and Cinful-1 (Llorens et al. 2011), also contained regions between the end of pol and the 3′-LTR but none with detectable vestiges of ORFs. However, there is a family of soybean elements within the Ogre lineage that, despite its close evolutionary relationship to other legume Ogre families that have no detectable env-like coding regions (Neumann et al. 2003; Macas and Neumann 2007), possesses an env-like 3′ eORF. GmOgre/SNARE is a family from G. max that shares the unusual features of Ogre lineage members—a conserved, intact 5′ eORF upstream of gag, a conserved intron in pol, and a minisatellite repeat region adjacent to the 3′-LTR (Laten et al. 2009; Du et al. 2010a). It is the most abundant transposon family in the soybean genome (Du et al. 2010b). But unlike all other members of the Ogre lineage, a GmOgre/SNARE consensus sequence from the end of pol to the minisatellite repeats contains an intact, 425-codon ORF whose conceptual translation generates a hypothetical protein with patches of significant similarity to the ENV-like hypothetical proteins from Cyclops-2 and Endovir1 (Laten et al. 2009). tBLASTn searches identified homologous coding regions in M. truncatula and L. japonicus in disrupted ORFs (Laten et al. 2009). What makes the GmOgre/SNARE ENV protein especially intriguing is the fact that Cyclops-2 is a member of the Ty3/Gypsy superfamily and Endovir1 is a member of the Ty1/Copia superfamily. This suggests that the ENV-like protein in GmOgre/SNARE may be a chimera. Du et al. (2010a) suggested that the GmOgre/SNARE env-like region represents a relatively recent capture event, but it also may reflect the maintenance of selective pressure in the G. max lineage and the relaxation of this pressure in the other lineages.
6.4 Origin of Plant env-Like Genes
Because of highly disrupted ORFs and the great diversity of conceptually translated env-like sequences, even from intact ORFs, homology that extends beyond closely related families, let alone to functionally characterized envelope proteins, is difficult to infer. Nor have these sequences been shown unequivocally to be homologous to any other characterized genes in plant or viral genomes. Nonetheless, it has been proposed and widely presumed that env-like coding regions were independently acquired or captured (from an unknown source or sources) by ancestral Ty1/Copia and/or Ty3/Gypsy retrotransposons (Peterson-Burch et al. 2000; Du et al. 2010b). The putative chimeric env-like region or GmOgre/SNARE might represent a more recent fusion event (Laten et al. 2009; Du et al. 2010b). A less likely but not inconceivable scenario is the possibility that some and perhaps many retrotransposons are actually the descendants of ancient enveloped retroviruses (Eickbush and Jamburuthugoda 2008) and that genomes, including those of plants (Yano et al. 2005), have recorded the history of the demise of env genes.
Based on a multi-sequence alignment of an unprecedentedly broad range of ENV sequences, Du et al. (2010b) created a neighbor joining tree linking sequences from plant Ty1/Copia and Ty3/Gypsy retroelements rooted to the Drosophila 17.6 ENV protein (Fig. 6.8). Conservation of ENV sequences between the superfamilies in the alignment is limited to a small number of identical residues and a larger number that are similar. But these similarities could also reflect convergent evolution and not evolutionary homology. Nonetheless, assuming homology, the Ty1/Copia sequences appeared to be monophyletic but the Ty3/Gypsy sequences were not. Instead, one clade of ENV sequences from Ty3/Gypsy elements in soybean, Lotus, and Medicago was the sister group to a subset of ENV sequences associated with elements belonging to the Ty1/Copia superfamily. The neighbor joining trees of the corresponding RT sequences did not generate this tree topology and conformed to the expected segregation of all members of the two superfamilies into two sister clades (Du et al. 2010b). The authors inferred that the Ty1/Copia env-like gene was acquired from an ancestral member of its sister Ty3/Gypsy clade, long after the capture of the env-like sequence by an ancestral Ty3/Gypsy retrotransposon near the crown of the tree (Fig. 6.8). However, this conclusion was based, in part, on the questionable rooting of the tree to the ENV sequence of a Drosophila element. Removal of the root generates an unrooted tree whose topology leaves open the question of the origin of the ENV sequences.
Neighbor joining tree generated from plant retroelement ENV-like sequences. Double asterisk represent nodes with 86–100 % bootstrap support; asterisk represent nodes with 64–75 % bootstrap support. Rooted to the putative Env protein from the Gypsy-like element, 17.6, in D. melanogaster. From Du et al. (2010b)
6.5 Function of Plant ENV Hypothetical Proteins
There can be little dispute that large numbers of plant retroelement families have possessed genes encoding transmembrane proteins sometime during their evolutionary history, and that in a few cases what have been called env-like genes still encode what appear to be potentially functional proteins. However, it seems unlikely that the expression of an env-like ORF was essential to the proliferation of most families in the Athila and Tat clades, although traces of their widespread distribution suggests an important function, even if that function was transient. The presence of highly conserved, intact env-like ORFs in the hundreds of copies of Sireviruses in G. max and L. japonicus could be due to strong selection or to their recent explosive amplification. One can only speculate whether those env-like genes that appear to have retained function are the products of continuing, lineage-specific, purifying selection, or resurrected Phoenixes that have emerged from the ashes of degenerate copies by a variety of mutational processes.
The possible function of plant retroelement ENV-like proteins has been the subject of much speculation in the nearly total absence of experimental data (Kumar 1998; Laten et al. 1998; Wright and Voytas 1998, 2002; Peterson-Burch et al. 2000; Vicient et al. 2001; Grandbastien 2008). Based on predicted secondary structural elements, and the suggested parallels to endogenous retroviruses in mammals and invertebrates, membrane fusion has been the most promoted candidate.
Membrane fusion might be an unlikely choice, however, since cell walls would preclude this mechanism as an efficient mode of transmission and systemic infection in plants. Most plant viruses are transmitted by insect vectors in which the viruses do not propagate in their insect hosts (Lazarowitz 2007). But in the case of a few, the viruses also infect the cells of their hosts (propagative viruses) and could just as well be considered animal viruses (Lazarowitz 2007). This latter group includes members of two families of enveloped viruses: Rhabdoviridae and Bunyaviridae. The former includes Sonchus Yellow Net Virus (SYNV) that generates a virion composed of a lipid envelope embedded with virally encoded glycoproteins, while the latter includes tospoviruses like Tomato Spotted Wilt Virus (TSWV) with a genome that encodes two envelope glycoproteins (Lazarowitz 2007; Whitfield et al. 2005). In their plant hosts, intracellular SYNV and TSWV particles appear to associate with the nuclear and ER membranes, respectively (Lazarowitz 2007). In the case of TSWV single-enveloped particles are formed and transferred to feeding thrips (Kikkert et al. 1999). In thrip hosts, TSWV virions are associated with the plasma membrane and are released from infected cells by fusion with the cell membrane (Whitfield et al. 2005). However, there are no reports of detected homology between any plant retroelement ENV-like hypothetical protein and those of plant enveloped viruses.
The maintenance of envelope-encoding sequences in these viruses appears to be directly related to infectivity in their animal hosts, not in their plant hosts. When maintained solely by serial mechanical inoculations from one infected plant to another, non-enveloped mutant isolates accumulate (Goldbach and Peters 1996). These isolates are fully capable of mounting a systemic infection in plants after mechanical transfer (Goldbach and Peters 1996). However, non-enveloped isolates with mutations in the glycoprotein genes have been shown to be incapable of reinfecting the thrip host (Nagata et al. 2000). These observations provide an attractive, albeit highly speculative, model for the existence of endogenous retrovirus lineages in plants with nonfunctional and functional env-like genes. Confirming this model would require at a minimum the discovery of related elements in invertebrate vectors and demonstrating that virions from plants could fuse with the plasma membranes of the invertebrate host. Attempts to detect SIRE1 using PCR amplification in several known vectors including several species of thrips and aphids were unsuccessful (Laten, unpublished). Nor have tBLASTn or BLASTn searches of the Genbank database retrieved any animal DNA or mRNA with significant similarity to plant env-like genes. (Laten, unpublished).
6.6 Concluding Remarks
While much is now known about the structure and evolutionary relationships of the large collection of plant retroelements in both the Ty1/Copia and Ty3/Gypsy superfamilies that possess a “mysterious” 3′ eORF downstream of pol, hard evidence for the function(s) of the encoded protein(s) remains elusive. Regardless of whether or not transcripts, spliced or otherwise, represent functional expression, no reports of protein products have been published, let alone the results of functional assays. Potentially functional ENV-like proteins need to be isolated, either from plant tissue or from cloned constructs. Assays need to be developed and optimized for the evaluation of not only putative functions, e.g., membrane fusion, but also for alternative functions. Viral envelope proteins are just one of the many classes of proteins characterized by transmembrane and/or coiled coil domains, although the model set by the structure and evolution of animal endogenous retroviruses has greatly influenced the annotations of these elements. Continuing to annotate as “env-like” 3′ eORFs whose conceptual translations produce hypothetical proteins with transmembrane domains seems ill-advised at the present time, and the question of the existence of plant retroviruses, endogenous or infectious, remains unanswered. Function notwithstanding, the env-like genes in plant genomes are arguably the most abundant protein coding regions in the genomes of higher plants for which no function has been determined.
References
Babu MM, Iyer LM, Balaji S, Aravind L (2006) The natural history of the WRKY-GCM1 zinc fingers and the relationship between transcription factors and transposons. Nucleic Acids Res 34:6505–6520
Bannert N, Kurth R (2006) The evolutionary dynamics of human endogenous retroviral families. Annu Rev Genom Hum Genet 7:149–173
Baucom RS, Estill JC, Chaparro C, Upshaw N, Jogi A, Deragon JM, Westerman RP, SanMiguel PJ, Bennetzen JL (2009) Exceptional diversity, non-random distribution, and rapid evolution of retroelements in the B73 maize genome. PLoS Genet 5:e1000732
Belshaw R, Pereira V, Katzourakis A, Talbot G, Paces J, Burt A, Tristem M (2004) Long-term reinfection of the human genome by endogenous retroviruses. Proc Natl Acad Sci USA 101:4894–4899
Belshaw R, Katzourakis A, Paces J, Burt A, Tristem M (2005) High copy number in human endogenous retrovirus families is associated with copying mechanisms in addition to reinfection. Mol Biol Evol 22:814–817
Benit L, Dessen P, Heidmann T (2001) Identification, phylogeny, and evolution of retroviral elements based on their envelope genes. J Virol 75:11709–11719
Boeke JD, Stoye JP (1997) Retrotransposons, endogenous retroviruses, and the evolution of retroelements. In: Coffin JM, Hughes SH, Varmus HE (eds) Retroviruses. Cold Spring Harbor Laboratory Press, Plainview, NY, pp 343–435
Boeke JD, Eickbush TH, Sandmeyer SB, Voytas DF (2005a) Family Metaviridae. In: Fauquet CM, Mayo MA, Maniloff J, Desselberger U, Ball LA (eds) Virus taxonomy: VIIIth report of the International Committee on Taxonomy of Viruses. Elsevier Academic, San Diego, CA, pp 409–420
Boeke JD, Eickbush TH, Sandmeyer SB, Voytas DF (2005b) Family Pseudoviridae. In: Fauquet CM, Mayo MA, Maniloff J, Desselberger U, Ball LA (eds) Virus taxonomy: VIIIth report of the International Committee on Taxonomy of Viruses. Elsevier Academic, San Diego, CA, pp 397–407
Bousios A, Darzentas N, Tsaftaris A, Pearce SR (2010) Highly conserved motifs in non-coding regions of Sirevirus retrotransposons: the key for their pattern of distribution within and across plants? BMC Genomics 11:89
Bousios A, Kourmpetis YAI, Pavlidis P, Minga E, Tsaftaris A, Darzentas N (2012a) The turbulent life of Sirevirus retrotransposons and the evolution of the maize genome: more than ten thousand elements tell the story. Plant J 69:475–488
Bousios A, Minga E, Kalitsou N, Pantermali M, Tsaballa A, Darzentas N (2012b) MASiVEdb: the Sirevirus plant retrotransposon database. BMC Genomics 13:158
Bowen NJ, McDonald JF (1999) Genomic analysis of Caenorhabditis elegans reveals ancient families of retroviral-like elements. Genome Res 9:924–935
Caffrey M (2011) HIV envelope: challenges and opportunities for development of entry inhibitors. Trends Microbiol 19:191–197
Chalvet F, Teysset L, Terzian C, Prud’homme N, Santamaria P, Bucheton A, Pelisson A (1999) Proviral amplification of the Gypsy endogenous retrovirus of Drosophila melanogaster involves env-independent invasion of the female germline. EMBO J 18:2659–2669
Chavanne F, Zhang DX, Liaud MF, Cerff R (1998) Structure and evolution of Cyclops: a novel giant retrotransposon of the Ty3/Gypsy family highly amplified in pea and other legume species. Plant Mol Biol 37:363–375
Cosset FL, Lavillette D (2011) Cell entry of enveloped viruses. Adv Genet 73:121–183
Daraselia ND, Tarchevskaya S, Narita JO (1996) The promoter for tomato 3-hydroxy-3-methylglutaryl coenzyme A reductase gene 2 has unusual regulatory elements that direct high-level expression. Plant Physiol 112:727–733
Du C, Swigonova Z, Messing J (2006) Retrotranspositions in orthologous regions of closely related grass species. BMC Evol Biol 6:62
Du J, Tian Z, Bowen NJ, Schmutz J, Shoemaker RC, Ma J (2010a) Bifurcation and enhancement of autonomous-nonautonomous retrotransposon partnership through LTR swapping in soybean. Plant Cell 22:48–61
Du J, Tian Z, Hans CS, Laten HM, Cannon SB, Jackson SA, Shoemaker RC, Ma J (2010b) Evolutionary conservation, diversity and specificity of LTR-retrotransposons in flowering plants: insights from genome-wide analysis and multi-specific comparison. Plant J 63:584–598
Eickbush TH, Jamburuthugoda VK (2008) The diversity of retrotransposons and the properties of their reverse transcriptases. Virus Res 134:221–234
Eickbush TH, Malik HS (2002) Origins and evolution of retrotransposons. In: Craig NL, Craigie R, Gellert M, Lambowitz AM (eds) Mobile DNA II. ASM Press, Washington, pp 1111–1144
Felder H, Herzceg A, de Chastonay Y, Aeby P, Tobler H, Muller F (1994) Tas, a retrotransposon from the parasitic nematode Ascaris lumbricoides. Gene 149:219–225
Feschotte C, Gilbert C (2012) Endogenous viruses: insights into viral evolution and impact on host biology. Nat Rev Genet 13:283–296
Finlay BB, McFadden G (2006) Anti-immunology: evasion of the host immune system by bacterial and viral pathogens. Cell 124:767–782
Friesen PD, Nissen MS (1990) Gene organization and transcription of TED, a lepidopteran retrotransposon integrated within the Baculovirus genome. Mol Cell Biol 10:3067–3077
Gaston GD (2011) Detection of SIRE1 ENV, a potential retroviral like protein in soybean. M.S. Thesis, Loyola University, Chicago
Gladyshev EA, Meselson M, Arkhipova IR (2007) A deep-branching clade of retrovirus-like retrotransposons in bdelloid rotifers. Gene 390:136–145
Goldbach R, Peters D (1996) Molecular and biological aspects of tospoviruses. In: Elliot RM (ed) The Bunyaviridae. Plenum Press, New York, pp 129–157
Gonzalez VM, Benjak A, Henaff EM, Mir G, Casacuberta JM, Garcia-Mas J, Puigdomenech P (2010) Sequencing of 6.7 Mb of the melon genome using a BAC pooling strategy. BMC Plant Biol 10:246
Grandbastien MA (2008) Retrotransposons in plants. In: Mahy BWJ, Van Regenmortel MHV (eds) Encyclopedia of plants. Elsevier, Oxford, pp 428–436
Hafez EE, Abdel Ghany AA, Paterson AH, Zaki EA (2009) Sequence heterogeneity of the envelope-like domain in cultivated allotetraploid Gossypium species and their diploid progenitors. J Appl Genet 50:17–23
Harrison SC (2008) Viral membrane fusion. Nat Struct Mol Biol 15:690–698
Havecker ER, Voytas DF (2003) The soybean retroelement SIRE1 uses stop codon suppression to express its envelope-like protein. EMBO Rep 4:274–277
Havecker ER, Gao X, Voytas DF (2005) The Sireviruses, a plant-specific lineage of the Ty1/copia retrotransposons, interact with a family of proteins related to dynein light chain 8. Plant Physiol 139:857–868
Hohn T, Richert-Poggeler KR, Staginnus C, Harper G, Schwarzacher T, Teo CH, Teycheney P-Y, Iskra-Caruana M-L, Hull R (2008) Evolution of integrated plant viruses. In: Roossinck MJ (ed) Plant virus evolution. Springer, Berlin, pp 53–81
Holligan D, Zhang X, Jiang N, Pritham EJ, Wessler SR (2006) The transposable element landscape of the model legume Lotus japonicus. Genetics 174:2215–2228
Howard G, Eiges R, Gaudet F, Jaenisch R, Eden A (2008) Activation and transposition of endogenous retroviral elements in hypomethylation induced tumors in mice. Oncogene 27:404–408
Hribova E, Neumann P, Matsumoto T, Roux N, Macas J, Dolezel J (2010) Repetitive part of the banana (Musa acuminata) genome investigated by low-depth 454 sequencing. BMC Plant Biol 10:204
Hunter E (1997) Viral entry and receptors. In: Coffin JM, Hughes SH, Varmus HE (eds) Retroviruses. Cold Spring Harbor Laboratory Press, Plainview, NY, pp 71–119
Jern P, Coffin JM (2008) Effects of retroviruses on host genome function. Annu Rev Genet 42:709–732
Jin YK, Bennetzen JL (1994) Integration and nonrandom mutation of a plasma membrane proton ATPase gene fragment within the Bs1 retroelement of maize. Plant Cell 6:1177–1186
Kadlec J, Loureiro S, Abrescia NGA, Stuart DI, Jones IM (2008) The post-fusion structure of Baculovirus gp64 supports a unified view of viral fusion machines. Nat Struct Mol Biol 15:1024–1030
Kapitonov VV, Jurka J (1999) Molecular paleontology of transposable elements from Arabidopsis thaliana. Genetica 107:27–37
Kenward KD, Bai D, Ban MR, Brandle JE (1999) Isolation and characterization of Tnd-1, a retrotransposon marker linked to black root rot resistance in tobacco. Theor Appl Genet 98:387–395
Kikkert M, van Lent J, Storms M, Bodegom P, Kormelink R, Goldbach R (1999) Tomato spotted wilt virus particle morphogenesis in plant cells. J Virol 73:2288–2297
Kim A, Terzian C, Santamaria P, Pelisson A, Prud’homme N, Bucheton A (1994) Retroviruses in invertebrates: the gypsy retrotransposon is apparently an infectious retrovirus of Drosophila melanogaster. Proc Natl Acad Sci USA 91:1285–1289
Kim FJ, Battini JL, Manel N, Sitbon M (2004) Emergence of vertebrate retroviruses and envelope capture. Virology 318:183–191
King AMQ, Adams MJ, Carstens EB, Lefkowitz EJ (eds) (2012) Virus taxonomy: ninth report of the international committee on taxonomy of viruses. Elsevier, London
Kumar A (1998) The evolution of plant retroviruses: moving to green pastures. Trends Plant Sci 3:371–374
Laten HM (1999) Phylogenetic evidence for Ty1-copia-like endogenous retroviruses in plant genomes. Genetica 107:87–93
Laten HM, Bousios A (2012) Genus Sirevirus. In: Tidona C, Darai G (eds) The Springer index of viruses, 2nd edn. Springer, New York, NY, pp 1561–1564
Laten HM, Morris RO (1993) SIRE-1, a long interspersed repetitive DNA element from soybean with weak sequence similarity to retrotransposons: initial characterization and partial sequence. Gene 134:153–159
Laten HM, Majumdar A, Gaucher EA (1998) SIRE-1, a copia/Ty1-like retroelement from soybean, encodes a retroviral envelope-like protein. Proc Natl Acad Sci USA 95:6897–6902
Laten HM, Havecker ER, Farmer LM, Voytas DF (2003) SIRE1, an endogenous retrovirus family from Glycine max, is highly homogeneous and evolutionarily young. Mol Biol Evol 20:1222–1230
Laten HM, Mogil LS, Wright LN (2009) A shotgun approach to discovering and reconstructing consensus retrotransposons ex novo from dense contigs of short sequences derived from Genbank Genome Survey Sequence database records. Gene 448:168–173
Lavin M, Herendeen PS, Wojciechowski MF (2005) Evolutionary rates analysis of Leguminosae implicates a rapid diversification of lineages during the tertiary. Syst Biol 54:575–594
Lazarowitz SD (2007) Plant viruses. In: Knipe DM, Howley PM (eds) Fields virology. Lippincott Williams & Wilkins, Philadelphia, PA, pp 641–705
Lerat E, Capy P (1999) Retrotransposons and retroviruses: analysis of the envelope gene. Mol Biol Evol 16:1198–1207
Li F, Ding S-W (2006) Virus counter-defense: diverse strategies for evading the RNA-silencing immunity. Annu Rev Microbiol 60:503–531
Lin E (2001) Analysis of SIRE1 transcriptional activity. M.S. Thesis, Loyola University, Chicago
Llorens JV, Clark JB, Martinez-Garay I, Soriano S, de Frutos R, Martinez-Sebastian MJ (2008) Gypsy endogenous retrovirus maintains potential infectivity in several species of Drosophilids. BMC Evol Biol 8:302
Llorens C, Muñoz-Pomer A, Bernad L, Botella H, Moya A (2009) Network dynamics of eukaryotic LTR retroelements beyond phylogenetic trees. Biol Direct 4:41
Llorens C, Futami R, Covelli L, Domínguez-Escribá L, Viu JM, Tamarit D, Aguilar-Rodríguez J, Vicente-Ripolles M, Fuster G, Bernet GP, Maumus F, Munoz-Pomer A, Sempere JM, Latorre A, Moya A (2011) The Gypsy database (GyDB) of mobile genetic elements: release 2.0. Nucleic Acids Res 39:D70–D74
Macas J, Neumann P (2007) Ogre elements–a distinct group of plant Ty3/gypsy-like retrotransposons. Gene 390:108–116
Macas J, Neumann P, Navratilova A (2007) Repetitive DNA in the pea (Pisum sativum L.) genome: comprehensive characterization using 454 sequencing and comparison to soybean and Medicago truncatula. BMC Genomics 8:427
Maksakova IA, Romanish MT, Gagnier L, Dunn CA, van de Lagemaat LN, Mager DL (2006) Retroviral elements and their hosts: insertional mutagenesis in the mouse germ line. PLoS Genet 2:e2
Maksakova IA, Mager DL, Reiss D (2008) Keeping active endogenous retroviral-like elements in check: the epigenetic perspective. Cell Mol Life Sci 65:3329–3347
Malik HS, Henikoff S, Eickbush TH (2000) Poised for contagion: evolutionary origins of the infectious abilities of invertebrate retroviruses. Genome Res 10:1307–1318
McCarthy EM, Liu J, Lizhi G, McDonald JF (2002) Long terminal repeat retrotransposons of Oryza sativa. Genome Biol 3:0053
Mejlumian L, Pelisson A, Bucheton A, Terzian C (2002) Comparative and functional studies of Drosophila species invasion by the gypsy endogenous retrovirus. Genetics 160:201–209
Misseri Y, Labesse G, Bucheton A, Terzian C (2003) Comparative sequence analysis and predictions for the envelope glycoproteins of insect endogenous retroviruses. Trends Microbiol 11:253–256
Misseri Y, Cerutti M, Devauchelle G, Bucheton A, Terzian C (2004) Analysis of the Drosophila gypsy endogenous retrovirus envelope glycoprotein. J Gen Virol 85:11–31
Moyes D, Griffiths DJ, Venables PJ (2007) Insertional polymorphisms: a new lease of life for endogenous retroviruses in human disease. Trends Genet 23:326–333
Nagaki K, Song J, Stupar RM, Parokonny AS, Yuan Q, Ouyang S, Liu J, Hsiao J, Jones KM, Dawe RK, Buell CR, Jiang J (2003) Molecular and cytological analyses of large tracks of centromeric DNA reveal the structure and evolutionary dynamics of maize centromeres. Genetics 163:759–770
Nagata T, Inoue-Nagata AK, Prins M, Goldbach R, Peters D (2000) Impeded thrips transmission of defective tomato spotted wilt virus isolates. Phytopathology 90:454–459
Neumann P, Pozarkova D, Macas J (2003) Highly abundant pea LTR retrotransposon Ogre is constitutively transcribed and partially spliced. Plant Mol Biol 53:399–410
Neumann P, Pozarkova D, Koblizkova A, Macas J (2005) PIGY, a new plant envelope-class LTR retrotransposon. Mol Genet Genomics 273:43–53
Nielen S, Campos-Fonseca F, Leal-Bertioli S, Guimaraes P, Seijo G, Town C, Arrial R, Bertioli D (2010) FIDEL-a retrovirus-like retrotransposon and its distinct evolutionary histories in the A- and B-genome components of cultivated peanut. Chromosome Res 18:227–246
Nishimura MT, Dangl JL (2010) Arabidopsis and the plant immune system. Plant J 61:1053–1066
Obbard DJ, Gordon KHJ, Buck AH, Jiggins FM (2009) The evolution of RNAi as a defence against viruses and transposable elements. Philos Trans R Soc Lond B Biol Sci 364:99–115
Ozers MS, Friesen PD (1996) The Env-like open reading frame of the Baculovirus-integrated retrotransposon TED encodes a retrovirus-like envelope protein. Virology 226:252–259
Pearce SR (2007) SIRE-1, a putative plant retrovirus is closely related to a legume TY1-copia retrotransposon family. Cell Mol Biol Lett 12:120–126
Pearson MN, Rohrmann GF (2002) Transfer, incorporation, and substitution of envelope fusion proteins among members of the Baculoviridae, Orthomyxoviridae, and Metaviridae (insect retrovirus) families. J Virol 76:5301–5304
Pearson MN, Rohrmann GF (2004) Conservation of a proteinase cleavage site between an insect retrovirus (gypsy) Env protein and a Baculovirus envelope fusion protein. Virology 322:61–68
Pearson MN, Rohrmann GF (2006) Envelope gene capture and insect retrovirus evolution: the relationship between Errantivirus and Baculovirus envelope proteins. Virus Res 118:7–15
Pelisson A, Mejlumian L, Robert V, Terzian C, Bucheton A (2002) Drosophila germline invasion by the endogenous retrovirus gypsy: involvement of the viral env gene. Insect Biochem Mol Biol 32:1249–1256
Peterson-Burch BD, Voytas DF (2002) Genes of the Pseudoviridae (Ty1/copia Retrotransposons). Mol Biol Evol 19:1832–1845
Peterson-Burch BD, Wright DA, Laten HM, Voytas DF (2000) Retroviruses in plants? Trends Genet 16:151–152
Piedöel M, Gonçalves IR, Higuet D, Bonnivard E (2011) Eukaryotic DIRS1-like retrotransposons: an overview. BMC Genomics 12:621
Pinter A, Honnen WJ (1988) O-linked glycosylation of retroviral envelope gene products. J Virol 62:1016–1021
Poulter R, Goodwin T (2005) DIRS-1 and the other tyrosine recombinase retrotransposons. Cytogenet Genome Res 110:575–588
Rabson AB, Graves BJ (1997) Synthesis and processing of viral RNA. In: Coffin JM, Hughes SH, Varmus HE (eds) Retroviruses. Cold Spring Harbor Laboratory Press, Plainview, NY, pp 205–261
Ramakrishna W, Dubcovsky J, Park YJ, Busso C, Emberton J, SanMiguel P, Bennetzen JL (2002) Different types and rates of genome evolution detected by comparative sequence analysis of orthologous segments from four cereal genomes. Genetics 162:1389–1400
Ribet D, Harper F, Dewannieux M, Pierron G, Heidmann T (2007) Murine MusD retrotransposon: structure and molecular evolution of an “intracellularized” retrovirus. J Virol 81:1888–1898
Ribet D, Harper F, Dupressoir A, Dewannieux M, Pierron G, Heidmann T (2008) An infectious progenitor for the murine IAP retrotransposon: emergence of an intracellular genetic parasite from an ancient retrovirus. Genome Res 18:597–609
Rohrmann GF, Karplus PA (2001) Relatedness of Baculovirus and gypsy retrotransposon envelope proteins. BMC Evol Biol 1:1
SanMiguel P, Vitte C (2009) The LTR-retrotransposons of maize. In: Bennetzen JL, Hake SC (eds) Handbook of maize: genetics and genomics. Springer, New York, NY, pp 307–327
SanMiguel P, Tikhonov A, Jin YK, Motchoulskaia N, Zakharov D, Melake-Berhan A, Springer PS, Edwards KJ, Lee M, Avramova Z, Bennetzen JL (1996) Nested retrotransposons in the intergenic regions of the maize genome. Science 274:765–768
Song SU, Gerasimova T, Kurkulos M, Boeke JD, Corces VG (1994) An env-like protein encoded by a Drosophila retroelement: evidence that gypsy is an infectious retrovirus. Genes Dev 8:2046–2057
Staginnus C, Richert-Poggeler KR (2006) Endogenous pararetroviruses: two-faced travelers in the plant genome. Trends Plant Sci 11:485–491
Steinbauerová V, Neumann P, Novák P, Macas J (2012) A widespread occurrence of extra open reading frames in plant Ty3/gypsy retrotransposons. Genetica. doi:10.1007/s10709-012-9654-9
Stocking C, Kozak CA (2008) Murine endogenous retroviruses. Cell Mol Life Sci 65:3383–3398
Terzian C, Pelisson A, Bucheton A (2001) Evolution and phylogeny of insect endogenous retroviruses. BMC Evol Biol 1:3
Teysset L, Burns JC, Shike H, Sullivan BL, Bucheton A, Terzian C (1998) A Moloney murine leukemia virus-based retroviral vector pseudotyped by the insect retroviral gypsy envelope can infect Drosophila cells. J Virol 72:853–856
Vicient CM, Kalendar R, Schulman AH (2001) Envelope-class retrovirus-like elements are widespread, transcribed and spliced, and insertionally polymorphic in plants. Genome Res 11:2041–2049
Vitte C, Bennetzen JL (2006) Analysis of retrotransposon structural diversity uncovers properties and propensities in angiosperm genome evolution. Proc Natl Acad Sci USA 103:17638–17643
Wang Y, Liska F, Gosele C, Sedova L, Kren V, Krenova D, Ivics Z, Hubner N, Izsvak Z (2010) A novel active endogenous retrovirus family contributes to genome variability in rat inbred strains. Genome Res 20:19–27
Wang X, Wang H, Wang J, Sun R, Wu J, Liu S, Bai Y, Mun JH, Bancroft I, Cheng F, Huang S, Li X, Hua W, Wang J, Wang X, Freeling M, Pires JC, Paterson AH, Chalhoub B, Wang B, Hayward A, Sharpe AG (2011) The genome of the mesopolyploid crop species Brassica rapa. Nat Genet 43:1035–1039
Weber B, Wenke T, Frommel U, Schmidt T, Heitkam T (2010) The Ty1-copia families SALIRE and Cotzilla populating the Beta vulgaris genome show remarkable differences in abundance, chromosomal distribution, and age. Chromosome Res 18:247–263
Whitfield AE, Ullman DE, German TL (2005) Tospovirus-thrips interactions. Annu Rev Phytopathol 43:459–489
Wicker T, Keller B (2007) Genome-wide comparative analysis of copia retrotransposons in Triticeae, rice, and Arabidopsis reveals conserved ancient evolutionary lineages and distinct dynamics of individual copia families. Genome Res 17:1072–1081
Wilson IB, Gavel Y, von Heijne G (1991) Amino acid distributions around O-linked glycosylation sites. Biochem J 275:529–534
Wright DA, Voytas DF (1998) Potential retroviruses in plants: Tat1 is related to a group of Arabidopsis thaliana Ty3/gypsy retrotransposons that encode envelope-like proteins. Genetics 149:703–715
Wright DA, Voytas DF (2002) Athila4 of Arabidopsis and Calypso of soybean define a lineage of endogenous plant retroviruses. Genome Res 12:122–131
Wu BW, Cannon PM, Gordon EM, Hall FL, Anderson WF (1998) Characterization of the proline-rich region of murine leukemia virus envelope protein. J Virol 72:5383–5391
Yang ZN, Ye XR, Molina J, Roose ML, Mirkov TE (2003) Sequence analysis of a 282-kilobase region surrounding the citrus Tristeza virus resistance gene (Ctv) locus in Poncirus trifoliata L. Raf. Plant Physiol 131:482–492
Yano ST, Panbehi B, Das A, Laten HM (2005) Diaspora, a large family of Ty3-gypsy retrotransposons in Glycine max, is an envelope-less member of an endogenous plant retrovirus lineage. BMC Evol Biol 5:30
Zhang Y, Maksakova IA, Gagnier L, van de Lagemaat LN, Mager DL (2008) Genome-wide assessments reveal extremely high levels of polymorphism of two active families of mouse endogenous retroviral elements. PLoS Genet 4:e1000007
Zhou Q, Haymer DS (1998) Molecular structure of yoyo, a gypsy-like retrotransposon from the Mediterranean fruit fly, Ceratitis capitata. Genetica 101:167–178
Author information
Authors and Affiliations
Corresponding author
Editor information
Editors and Affiliations
Rights and permissions
Copyright information
© 2012 Springer-Verlag Berlin Heidelberg
About this chapter
Cite this chapter
Laten, H.M., Gaston, G.D. (2012). Plant Endogenous Retroviruses? A Case of Mysterious ORFs. In: Grandbastien, MA., Casacuberta, J. (eds) Plant Transposable Elements. Topics in Current Genetics, vol 24. Springer, Berlin, Heidelberg. https://doi.org/10.1007/978-3-642-31842-9_6
Download citation
DOI: https://doi.org/10.1007/978-3-642-31842-9_6
Published:
Publisher Name: Springer, Berlin, Heidelberg
Print ISBN: 978-3-642-31841-2
Online ISBN: 978-3-642-31842-9
eBook Packages: Biomedical and Life SciencesBiomedical and Life Sciences (R0)