Abstract
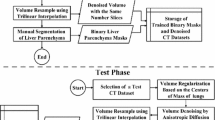
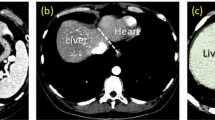
Modern epidemiological studies analyze a high amount of magnetic resonance imaging (MRI) data, which requires fully automatic segmentation methods to assist in organ volumetry. We propose a fully automatic two-step 3D level set algorithm for liver segmentation in MRI data that delineates liver tissue on liver probability maps and uses a distance transform based segmentation refinement method to improve segmentation results. MR intensity distributions in test subjects are extracted in a training phase to obtain prior information on liver, kidney and background tissue types. Probability maps are generated by using linear discriminant analysis and Bayesian methods. The algorithm is able to differentiate between normal liver tissue and fatty liver tissue and generates probability maps for both tissues to improve the segmentation results. The algorithm is embedded in a volumetry framework and yields sufficiently good results for use in epidemiological studies.
Chapter PDF
Similar content being viewed by others
References
Gao, L., Heath, D., Kuszyk, B., Fishman, E.: Automatic liver segmentation technique for three-dimensional visualization of CT data. Radiology 201(2), 359–364 (1996)
Seo, K.S.: Improved Fully Automatic Liver Segmentation Using Histogram Tail Threshold Algorithms. In: International Conference on Computational Science, pp. 822–825 (2005)
Evans, A., Lambrou, T., Linnery, A., Todd-Pokroped, A.: Automatic Segmentation of Liver Using a Toplogy Adaptive Snake. In: International Conference on Biomedical Engineering, pp. 205–208 (2004)
Schenk, A., Prause, G.P.M., Peitgen, H.O.: Efficient semiautomatic segmentation of 3d objects in medical images. In: Medical Image Computing and Computer-Assisted Intervention, pp. 186–195 (2001)
Daisuke, F., Akinobu, S., Hidefumi, K.: Automatic Liver Segmentation Method based on Maximum A Posterior Probability Estimation and LevelSet Method. In: Medical Image Computing and Computer Assisted Intervention, pp. 117–124 (2007)
Lamecker, H., Lange, T., Seebass, M.: A Statistical Shape Model for the Liver. In: Dohi, T., Kikinis, R. (eds.) MICCAI 2002. LNCS, vol. 2489, pp. 421–427. Springer, Heidelberg (2002)
Ling, H., Zhou, S.K., Zheng, Y., Georgescu, B., Suehling, M., Comaniciu, D.: Hierarchical, learning-based automatic liver segmentation. In: IEEE Conf. on Computer Vision and Pattern Recognition (CVPR 2008). IEEE Computer Society, Anchorage (2008)
Soler, L., Delingette, H., Malandain, G., Montagnat, J., Ayache, N., Koehle, C., Dourthe, O., Malassagne, B., Smith, M., Mutter, D., et al.: Fully Automatic Anatomical, Pathological, and Functional Segmentation from CT Scans for Hepatic Surgery. In: Hanson, K.M. (ed.), February 14, pp. 246–255. SPIE, San Diego (2000)
Heimann, T., Wolf, I., Meinzer, H.-P.: Active shape models for a fully automated 3D segmentation of the liver – an evaluation on clinical data. In: Larsen, R., Nielsen, M., Sporring, J. (eds.) MICCAI 2006. LNCS, vol. 4191, pp. 41–48. Springer, Heidelberg (2006)
Li, M., Yang, L.: Liver Segmentation Based on Expectation Maximization and Morphological Filters in CT Images. In: Bioinformatics and Biomedical Engineering (ICBBE 2007). IEEE, Wuhan (2007)
Freiman, M., Eliassaf, O., Taieb, Y., Joskowicz, L., Azraq, Y., Sosna, J.: An iterative Bayesian approach for nearly automatic liver segmentation: algorithm and validation. International Journal of Computer Assisted Radiology and Surgery 3(5), 439–446 (2008)
Sosna, J., Berman, P., Azraq, Y., Libson, E.: Liver segmentation and volume calculation from MDCT using Bayesian likelihood maximization technique: comparison with manual tracing technique. RSNA, Chicago (2006)
Massoptier, L., Casciaro, S.: Fully automatic liver segmentation through graph-cut technique. In: Rousseau, J., Delhomme, G., Akay, M. (eds.), Lyon, France, August 22-26, pp. 5243–5246. IEEE, Los Alamitos (2007)
Beichel, R., Bauer, C., Bornik, A., Sorantin, E., Bischof, H.: Liver Segmentation in CT Data: A Segmentation Refinement Approach. In: Ayache, N., Ourselin, S., Maeder, A. (eds.), Brisbane, Australia, pp. 235–245. Springer, Heidelberg (October 29, 2007)
Rusko, L., Bekes, G.: Liver segmentation for contrast-enhanced MR images using partitioned probabilistic model. International Journal of Computer Assisted Radiology and Surgery 6(1), 13–20 (2010)
Platero, C., Gonzalez, M., Tobar, M.C., Poncela, J.M., Sanguino, J., Asensio, G., Santos, E.: Automatic method to segment the liver on multi-phase MRI. In: Jover, J.H. (ed.), Barcelona, Spain, June 25-28 (2008)
Cheng, K., Gu, L., Xu, J.: A novel shape prior based level set method for liver segmentation from MR Images, Shenzhen, China, May 30-31, pp. 144–147 (2008)
Chen, G., Gu, L., Qian, L., Xu, J.: An Improved Level Set for Liver Segmentation and Perfusion Analysis in MRIs. IEEE Transactions on Information Technology in Biomedicine 3(1), 94–103 (2009)
Yuan, Z., Wang, Y., Yang, J., Liu, Y.: A novel automatic liver segmentation technique for MR images. In: Image and Signal Processing (CISP), Yantai, pp. 1282–1286 (2010)
Baumeister, S.E., Voelzke, H., Marschall, P., John, U., Schmidt, C.O., Flessa, S., Alte, D.: Impact of Fatty Liver Disease on Health Care Utilization and Costs in a General Population: A 5-Year Observation. Gastroenterology 134(1), 85–94 (2008)
Caselles, V., Kimmel, R., Sapiro, G.: Geodesic active contours. In: International Conference on Computer Vision (ICCV 1995). IEEE Computer Society, Massachusetts (1995)
Hussain, H.K., Chenevert, T.L., Londy, F.J., Gulani, V., Swanson, S.D., McKenna, B.J., Appelman, H.D., Adusumilli, S., Greenson, J.K., Conjeevaram, H.: Hepatic fat fraction: MR imaging for quantitative measurement and display early experience. Radiology 237, 1048–1055 (2005)
Whitaker, R.T., Xue, X.: Variable-conductance, Level-Set Curvature for Image Denoising, Thessaloniki, Greece, pp. 142–145 (2001)
Gloger, O., Kuehn, J., Stanski, A., Voelzke, H., Puls, R.: A fully automatic three-step liver segmentation method on LDA-based probability maps for multiple contrast MR images. Magnetic Resonance Imaging 28(6), 882–897 (2010)
Fisher, R.: The statistical utilization of multiple measurements. Ann. Eugenics 8, 376–386 (1938)
Rao, C.: The utilization of multiple measurements in problems of biological classification. Journal of the Royal Statistical Society 10, 159–203 (1948)
Crum, W.R., Camara, O., Hill, D.L.G.: Generalized Overlap Measures for Evalution and Validation in Medical Image Analysis. IEEE Transactions on Medical Imaging 25(11), 1451–1458 (2006)
Maier, F., Wimmer, A., Soza, G., Kaftan, J.N., Fritz, D., Dillmann, R.: Automatic Liver Segmentation Using the Random Walker Algorithm. Bildverarbeitung fuer die Medizin, pp. 65–61 (2008)
Chan, T.F., Vese, L.A.: Active contours without edges. IEEE Transactions on Image Processing 10(2), 266–277 (2001)
Author information
Authors and Affiliations
Editor information
Editors and Affiliations
Rights and permissions
Copyright information
© 2011 Springer-Verlag Berlin Heidelberg
About this paper
Cite this paper
Gloger, O., Toennies, K., Kuehn, JP. (2011). Fully Automatic Liver Volumetry Using 3D Level Set Segmentation for Differentiated Liver Tissue Types in Multiple Contrast MR Datasets. In: Heyden, A., Kahl, F. (eds) Image Analysis. SCIA 2011. Lecture Notes in Computer Science, vol 6688. Springer, Berlin, Heidelberg. https://doi.org/10.1007/978-3-642-21227-7_48
Download citation
DOI: https://doi.org/10.1007/978-3-642-21227-7_48
Publisher Name: Springer, Berlin, Heidelberg
Print ISBN: 978-3-642-21226-0
Online ISBN: 978-3-642-21227-7
eBook Packages: Computer ScienceComputer Science (R0)