Abstract
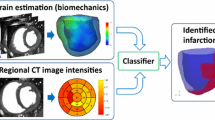
Different heart tissue identification is important for therapeutic decision-making in patients with myocardial infarction (MI), this provides physicians with a better clinical decision-making tool. Composite Strain Encoding (C-SENC) is an MRI acquisition technique that is used to acquire cardiac tissue viability and contractility images. It combines the use of blackblood delayed-enhancement (DE) imaging to identify the infracted (dead) tissue inside the heart muscle and the ability to image myocardial deformation from the strain-encoding (SENC) imaging technique. In this work, various machine learning techniques are applied to identify the different heart tissues and the background regions in the C-SENC images. The proposed methods are tested using numerical simulations of the heart C-SENC images and real images of patients. The results show that the applied techniques are able to identify the different components of the image with a high accuracy.
Chapter PDF
Similar content being viewed by others
Keywords
These keywords were added by machine and not by the authors. This process is experimental and the keywords may be updated as the learning algorithm improves.
References
Ibrahim, E.-S.H., Stuber, M., Kraitchman, D.L., Weiss, R.G., Osman, N.F.: Combined Functional and Viability Cardiac MR Imaging in a Single Breathhold. Magn. Reson. Med. 58, 843–849 (2007)
Watzinger, N., Saeed, M., Wendland, M.F., Akbari, H., Lund, G., Higgins, C.B.: Myocardial viability: magnetic resonance assessment of functional reserve and tissue characterization. J. Cardiovasc. Magn. Reson. 3, 195–208 (2001)
Kim, R.J., Wu, E., Rafael, A., Chen, E., Parker, M.A., Simonetti, O., Klocke, F.J., Bonow, R.O., Judd, R.M.: The use of contrast-enhanced magnetic resonance imaging to identify reversible myocardial dysfunction. N. Engl. J. Med. 343, 1445–1453 (2000)
Osman, N.F., Sampath, S., Atalar, E., Prince, J.L.: Imaging longitudinal cardiac strain on short-axis images using strain-encoding MRI. Magn. Reson. Med. 46, 324–334 (2001)
Ibrahim, E.-S.H., Weiss, R.G., Stuber, M., Spooner, A.E., Osman, N.F.: Identification of Different Heart Tissues from MRI C-SENC Images Using an unsupervised Multi-Stage Fuzzy Clustering Technique. J. Magn. Reson. Imaging 28(2), 519–526 (2008)
Motaal, A.G., El Gayar, N., Osman, N.F.: Automated Cardiac-Tissue Identification in Composite Strain-Encoded (C-SENC) Images Using Fuzzy C-means and Bayesian Classifier. Accepted in 4th International Conference on Bioinformatics and Biomedical Engineering (iCBBE 2010), Chengdu, China (2010)
Sharma, P., Socolow, J., Patel, S., Pettigrew, R., Oshinski, J.: Effect of Gd-DTPA-BMA on blood and myocardial T1 at 1.5T and 3T in humans. Magn. Reson. Imaging 23, 323–330 (2006)
Witten, L.I., Frank, E.: Data Mining: Practical Machine Learning Tools and Techniques with Java Implmentations. Morgan Kaufmann, San Francisco (1999)
Author information
Authors and Affiliations
Editor information
Editors and Affiliations
Rights and permissions
Copyright information
© 2010 Springer-Verlag Berlin Heidelberg
About this paper
Cite this paper
Motaal, A.G., El-Gayar, N., Osman, N.F. (2010). Different Regions Identification in Composite Strain-Encoded (C-SENC) Images Using Machine Learning Techniques. In: Schwenker, F., El Gayar, N. (eds) Artificial Neural Networks in Pattern Recognition. ANNPR 2010. Lecture Notes in Computer Science(), vol 5998. Springer, Berlin, Heidelberg. https://doi.org/10.1007/978-3-642-12159-3_21
Download citation
DOI: https://doi.org/10.1007/978-3-642-12159-3_21
Publisher Name: Springer, Berlin, Heidelberg
Print ISBN: 978-3-642-12158-6
Online ISBN: 978-3-642-12159-3
eBook Packages: Computer ScienceComputer Science (R0)