Abstract
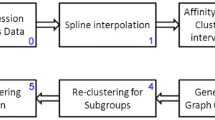
Genes with similar expression profiles are expected to be functionally related or co-regulated. In this direction, clustering microarray time-series data via pairwise alignment of piece-wise linear profiles has been recently introduced. We propose a k-means clustering approach based on a multiple alignment of natural cubic spline representations of gene expression profiles. The multiple alignment is achieved by minimizing the sum of integrated squared errors over a time-interval, defined on a set of profiles. Preliminary experiments on a well-known data set of 221 pre-clustered Saccharomyces cerevisiae gene expression profiles yields excellent results with 79.64% accuracy.
Chapter PDF
Similar content being viewed by others
Keywords
References
Cho, R., Campbell, M., Winzeler, E., Steinmetz, L., Conway, A., Wodicka, L., Wolfsberg, T., Gareilian, A., Lockhart, D., Davis, R.: A genome-wide transactional analysis of the mitotic cell cycle. Molecular Cell 2(1), 65–73 (1998)
Bar-Joseph, Z., Gerber, G., Jaakkola, T., Gifford, D., Simon, I.: Continuous representations of time series gene expresion data. Journal of Comp. Biology 10(3-4) (2003)
Bréhélin, L.: Clustering gene expression series with prior knowledge. In: Casadio, R., Myers, G. (eds.) WABI 2005. LNCS (LNBI), vol. 3692, pp. 27–38. Springer, Heidelberg (2005)
Chu, S., DeRisi, J., Eisen, M., Mulholland, J., Botstein, D., Brown, P., Herskowitz, I.: The transcriptional program of sporulation in budding yeast. Science 282, 699–705 (1998)
Djean, S., Martin, P., Baccini, A., Besse, P.: Clustering time-series gene expression data using smoothing spline derivatives. EURASIP Journal on Bioinformatics and Systems Biology 70561, 705–761 (2007)
Ernst, J., Nau, G., Bar-Joseph, Z.: Clustering short time series gene expression data. Bioinformatics 21(suppl. 1), i159–i168 (2005)
Ramoni, M., Sebastiani, P., Kohane, I. (eds.): Cluster analysis of gene expression dynamics. Proc. Natl. Acad. Sci. USA 99 (2002)
Tavazoie, S., Hughes, J., Campbell, M., Cho, R., Church, G.: Systematic determination of genetic network architecture. Nature Genetics 22, 281–285 (1999)
Tamayo, P., Slonim, D., Mesirov, J., Zhu, Q., Kitareewan, S., Dmitrovsky, E., Lander, E., Golub, T. (eds.): Interpreting patterns of gene expression with SOMs: Methods and application to hematopoietic differentiation, vol. 96 (1999)
Heyer, L., Kruglyak, S., Yooseph, S.: Exploring expression data: identification and analysis of coexpressed genes. Genome Research 9, 1106–1115 (1999)
Moller-Levet, C., Klawonn, F., Cho, K., Wolkenhauer, O.: Clustering of unevenly sampled gene expression time-series data. Fuzzy sets and Systems 152(1-16), 49–66 (2005)
Peddada, S., Lobenhofer, E., Li, L., Afshari, C., Weinberg, C., Umbach, D.: Gene selection and clustering for time-course and dose-response microarray experiments using order-restricted inference. Bioinformatics 19(7), 834–841 (2003)
Rueda, L., Bari, A., Ngom, A.: Clustering time-series gene expression data with unequal time intervals. In: Priami, C., Dressler, F., Akan, O.B., Ngom, A. (eds.) Transactions on Computational Systems Biology X. LNCS (LNBI), vol. 5410, pp. 100–123. Springer, Heidelberg (2008)
Xu, R., Wunsch, D.: Clustering. Wiley-IEEE Press, Chichester (2008)
Roth, V., Laub, J., Kawanabe, M., Buhmann, J.: Optimal cluster preserving embedding of nonmetric proximity data. IEEE Trans. on Pattern Analysis and Machine Intelligence 25(12), 1540–1551 (2003)
Kuhn, H.: The hungarian method for the assignment problem. Naval Research Logistics 52(1), 7–21 (2005)
Author information
Authors and Affiliations
Editor information
Editors and Affiliations
Rights and permissions
Copyright information
© 2009 Springer-Verlag Berlin Heidelberg
About this paper
Cite this paper
Subhani, N., Ngom, A., Rueda, L., Burden, C. (2009). Microarray Time-Series Data Clustering via Multiple Alignment of Gene Expression Profiles. In: Kadirkamanathan, V., Sanguinetti, G., Girolami, M., Niranjan, M., Noirel, J. (eds) Pattern Recognition in Bioinformatics. PRIB 2009. Lecture Notes in Computer Science(), vol 5780. Springer, Berlin, Heidelberg. https://doi.org/10.1007/978-3-642-04031-3_33
Download citation
DOI: https://doi.org/10.1007/978-3-642-04031-3_33
Publisher Name: Springer, Berlin, Heidelberg
Print ISBN: 978-3-642-04030-6
Online ISBN: 978-3-642-04031-3
eBook Packages: Computer ScienceComputer Science (R0)