Abstract
The SARS coronavirus (SARS-CoV) replicase gene encodes 16 nonstructural proteins (nsps) with multiple enzymatic activities. Several of these enzymes are common components of replication complexes of other plus-strand RNA viruses, such as picornavirus 3C-like protease, papain-like protease, RNA-dependent RNA polymerase, RNA helicase, and ribose 2′-O-methyltransferase activities, while others such as exoribonuclease, endoribonuclease, and adenosine diphosphate-ribose 1″-phosphatase activities, are rarely or not conserved in viruses outside the order Nidovirales. The latter enzymes are believed to be involved in unique metabolic pathways used by coronaviruses to (1) replicate and transcribe their extremely large RNA genomes, and (2) interfere with cellular functions and antiviral host responses.
Since the global outbreak of SARS in 2003, major efforts have been made to elucidate the structures of the protein components of the SARS-CoV replication/transcription complex. Thus, in less than 5 years, the structures of as many as 16 SARS-CoV proteins or functional domains have been determined. Remarkably, eight of these 16 structures had novel folds, illustrating the uniqueness of the coronavirus replicative machinery. Furthermore, several new protein functions and potential drug targets have been identified in these studies. Current structural studies mainly focus on the few remaining proteins for which no structural information is available and larger protein complexes comprised of different nsps. The studies aim at obtaining detailed information on the functions and macromolecular assembly of the coronavirus replication/transcription machinery which, over a long period of time, may be used to develop selective antiviral drugs. This chapter reviews structural information on the SARS-CoV macro domain (ADRP) as well as nsps 7, 8, 9, and 15 and summarizes our current knowledge of active-site residues and intermolecular interactions of these proteins.
You have full access to this open access chapter, Download chapter PDF
Similar content being viewed by others
Keywords
- Electrophoretic Mobility Shift Assay
- Rubella Virus
- Large Protein Complex
- Murine Hepatitis Virus
- Nsp10 Protein
These keywords were added by machine and not by the authors. This process is experimental and the keywords may be updated as the learning algorithm improves.
1 The ADP-Ribose-1″-Phosphatase Domain
The SARS coronavirus (SARS-CoV) ADP-ribose-1″-phosphatase (ADRP) is part of nonstructural protein nsp3, a large multidomain protein of 1,922 amino acids (Snijder et al. 2003; Thiel et al. 2003). The protein is thought to contain at least seven domains: (1) N-terminal Glu/Asp-rich domain (acidic domain, AD); (2) ADRP domain (also called macro domain or X domain); (3) SUD (for “SARS-CoV unique domain,” not conserved in other coronaviruses); (4) papain-like protease (PLpro), which cleaves the viral polyproteins at three N-proximal sites and also has deubiquitinating activity (Barretto et al. 2005; Harcourt et al. 2004; Lindner et al. 2005; Ratia et al. 2006; Thiel et al. 2003); (5) an uncharacterized domain possibly extending the papain-like protease domain (termed PLnc for papain-like noncanonical); (6) a transmembrane domain (Kanjanahaluethai et al. 2007) in the N-terminal region of the Y domain (Ziebuhr et al. 2001); and (7) the remainder of the Y domain which includes a number of conserved metal-binding residues.
Macro domains are conserved in a wide variety of bacteria, archaea and eukaryotes. The name macro domain refers to the early finding that members of this group of proteins are related to the nonhistone domain of the histone macroH2A (Pehrson and Fried 1992). Macro domains are also conserved in a number of positive-strand RNA viruses, including specific genera of the Nidovirales, members of the Togaviridae, Rubella virus and Hepatitis E virus (HEV) (Gorbalenya et al. 1991; Snijder et al. 2003). The biological role of macro domains in the life cycle of positive-strand RNA viruses has not been resolved. ADRP is a side-product of cellular pre-tRNA splicing, a pathway seemingly unrelated to viral RNA replication. ADRP activity was first identified for a macro domain homolog from yeast using a proteomics approach (Martzen et al. 1999) and subsequently demonstrated for other related proteins including homologs from three coronaviruses, HCoV-229E, TGEV, and SARS-CoV, as well as alphaviruses and HEV (Egloff et al. 2006; Putics et al. 2005, 2006). Inactivation of macro domain-associated ADRP activity in HCoV-229E did not affect viral genome replication and subgenomic RNA synthesis in cell culture, suggesting that the activity, which is conserved across members of the Coronaviridae, may have important functions in vivo, for example in evading antiviral host responses (Putics et al. 2005).
The structure of the SARS-CoV macro domain has been determined at 1.8 Å resolution, both in the apo form and in complex with ADP-ribose (Egloff et al. 2006; Saikatendu et al. 2005) (Fig. 7.1). To date, the catalytic residues of the coronavirus macro domain have not been well defined. Only one absolutely conserved Asn residue (N41, Fig. 7.1a) could be implicated in ADRP activity whereas other residues in the vicinity of the catalytic center are not conserved and therefore less likely to be critically involved in catalysis. Similar to archaebacterial macro domains, SARS-CoV and several other RNA virus macro domains were shown to have poly(ADP-ribose)-binding activities (Egloff et al. 2006). A possible binding mode for poly(ADP-ribose) which does not appear to involve a charged groove (Fig. 7.1b) has been modeled by Egloff et al. (2006). The significance of the observed poly(ADP-ribose)-binding activity and the biologically relevant substrates of RNA virus macro domains are currently not clear and remain to be studied in more detail.
Crystal structure of the SARS-CoV nsp3 macro domain (from Egloff et al. 2006; PDB 2FAV). (a) Ribbon representation of the SARS-CoV macro domain in complex with ADP-ribose. Shown is the only conserved residue whose mutation abrogates activity in all homologs studied so far (N41). (b) Surface potential analysis (blue, positive charge; red, negative charge) of the whole domain showing the presumed active site accommodating an ADP-ribose molecule. Images were generated using PYMOL
2 The nsp7–nsp8–nsp9–nsp10 Cistron
The nsp7 to nsp10 proteins are encoded by the 3′-terminal ORF1a region. The proteins are highly conserved amongst members of the genus Coronavirus and, albeit to a lesser extent, members of other genera within the Coronaviridae family. Interestingly, the proteins encoded in the equivalent 3′-terminal ORF1a region of arterivirus genomes do not appear to be closely related to the coronavirus nsp7 to nsp10 proteins (Pasternak et al. 2006), which may indicate differences in the structures, functions and subunit compositions of replication/transcription complexes between the various families within the order Nidovirales. Using a reverse genetics approach, nsp 7, 8, 9, and 10 were shown to be essential for replication of murine hepatitis virus (MHV) in cell culture (Deming et al. 2007). Deletion of any of the four proteins abolished RNA synthesis and, consequently, production of infectious virus progeny (Deming et al. 2007). The precise functions of the proteins in viral replication remain to be characterized.
2.1 The nsp7 Protein
Nsp7 is a small protein of about 10 kDa that is well conserved in coronaviruses but has no detectable homolog outside the Coronaviridae. The SARS-CoV nsp7 structure was solved by NMR (Peti et al. 2005). The protein features a single domain with a novel fold comprised of five helical secondary structures (Fig. 7.2a) whose mutual positions are stabilized by a number of interhelical side-chain interactions. The residues involved in these interactions are predominantly hydrophobic; they form two interdigitated layers that hold the helices together and thus stabilize the fold. The surface charge distribution is asymmetrical (Fig. 7.2b) and both surfaces may be involved in protein–protein interactions.
Views of the solution structure of SARS-CoV nsp7 (from Peti et al. 2005; PDB 1YSY). (a) Ribbon presentation of the SARS-CoV nsp7 NMR structure. (b) The surface shows the electrostatic potential (blue, positive charge; red, negative charge). Images were generated using PYMOL
In infected cells, coronavirus nsp7 and/or nsp7-containing precursors localize to cytoplasmic membrane structures which are thought to be the sites of viral replication (Bost et al. 2000; Ng et al. 2001). SARS-CoV nsp7 was shown to dimerize and interact with nsp5, nsp8 (see below), nsp9 and nsp13 (von Brunn et al. 2007; Zhai et al. 2005). Furthermore, the MHV nsp7 was shown to interact specifically with nsp1 and nsp10 (Brockway et al. 2004).
2.2 The nsp8 Protein
The protein has a molecular mass of about 22 kDa and is conserved among the various genera of the family Coronaviridae. It has interesting functional and structural properties. Initial crystallization trials of bacterially expressed SARS-CoV nsp8 remained unsuccessful until 2005, when the crystal structure of SARS-CoV nsp8 in complex with SARS-CoV nsp7 was solved (Zhai et al. 2005). The crystal structure revealed a hexadecameric complex composed of eight molecules of nsp8 and eight molecules of nsp7 (Fig. 7.3a). This so-called nsp7–nsp8 supercomplex has an intricate hollow cylindrical structure with an inner diameter of about 30 Å. Most of the nsp8 residues that face the interior of the channel are highly conserved among coronaviruses. The inner dimensions and electrostatic properties enable the cylindrical nsp7−nsp8 structure to encircle and interact with nucleic acid. Interactions with dsRNA were demonstrated for nsp8 (but not nsp7) by electrophoretic mobility shift assays and critical residues involved in RNA binding were identified by mutagenesis.
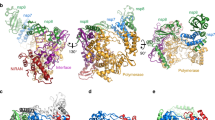
Different views of the SARS-CoV nsp7−nsp8 supercomplex (from Zhai et al. 2005; PDB 2AHM). (a) Structure of the nsp7−nsp8 hexadecamer supercomplex. Nsp7 and the two conformations of nsp8 are colored green, blue, and orange, respectively. (b) The two alternative nsp8 conformations. (c) Model of two nsp8 monomers in complex with a ssRNA template (5′-UAGC-3′) and two nucleotides (GTP and CTP). RNA template, GTP, and CTP are shown by a stick model. Two amino acid residues, Lys-58 and Arg-75, whose substitution abolished activity are represented in yellow. The two first NTPs incorporated (GTP in +1 and CTP in +2) are indicated. (d) The surface is colored according to electrostatic potential (blue, positive charge; red, negative charge). Images were generated using PYMOL
The α-helical fold seen in the previously reported NMR solution structure of nsp7 (Peti et al. 2005) was also seen in the crystal structure, where nsp7 was part of a complex with Nsp8. nsp8 was found to possess a novel fold with two slightly different conformations. One of the conformations was described as a “golf-club”-like structure composed of an N-terminal α-helical “shaft” domain and a C-terminal, mixed α/β “head” domain (Fig. 7.3b). The second conformation was described as resembling a golf club with a bent shaft, whereas the rest of the structure (particularly, the head domain) is very similar to the first structure (Fig. 7.3b). The eight nsp8 molecules constitute the framework of a large protein complex made up of “bricks” (nsp8) and “mortar” (nsp7), with nsp7 filling some of the remaining space between individual nsp8 molecules, thereby stabilizing the structure of the complex. Despite a number of intricate interactions between nsp7 and nsp8, the absence of nsp7 is not generally thought to markedly change the overall shape of the structure. On the basis of the architecture and electrostatic properties of the complex it was suggested that nsp7−nsp8 might function as a processivity factor, similar to bacterial (β2-clamp) or eukaryotic DNA polymerase processivity factors, such as PCNA (proliferating cell nuclear antigen).
This hypothesis was supported and significantly extended by data demonstrating that SARS-CoV nsp8 represents a second, noncanonical RNA-dependent RNA polymerase (RdRp) (Imbert et al. 2006). A recombinant form of the SARS-CoV nsp8 proved to be capable of synthesizing short oligonucleotides (<6 residues). It had relatively low fidelity and strongly preferred RNA with a 5′-(G/U)CC-3′ trinucleotide consensus sequence as a template. Three charged/polar residues, Lys-58, Lys-82 and Ser-85, all of them located in the N-terminal domain, were found to be essential for activity and may be part of a network of residues that catalyzes the phosphoryl transfer reaction. A structure and activity-based model of the nsp8-associated RdRp activity is presented in Fig. 7.3c,d. The structure of the C-terminal “head” domain of nsp8 appears to be distantly related to the catalytic palm subdomain of RNA virus RdRps. Moreover, it was shown that the “canonical” coronavirus RdRp domain residing in nsp12 contains a conserved sequence, called motif G, that is usually found in primer-dependent RNA polymerases (Gorbalenya et al. 2002). Taken together, the available information suggests that nsp8 may function as a primase to catalyze the synthesis of RNA primers to be used by the primer-dependent coronavirus nsp12 RdRp.
The presumed central role of nsp8 in coronavirus replication gains additional support by data suggesting that nsp8 interacts with a large number of SARS-CoV replicative proteins, including nsp2, nsp5 (3CLpro), nsp6, nsp7, nsp8, nsp9, nsp12 (RdRp), nsp13 (helicase), and nsp14 (exoribonuclease) (Imbert et al. 2008; von Brunn et al. 2007). Finally, an elegant MHV reverse genetics study provided evidence to suggest interactions between nsp8 (and also nsp9) with the 3′untranslated region, which is consistent with the proposed role of these proteins in coronavirus RNA synthesis, possibly the initiation of minus-strand RNA synthesis (Zust et al. 2008).
2.3 The nsp9 Protein
Crystal structures of nsp9, a protein of about 13 kDa, were solved simultaneously in two laboratories in 2004, one to a resolution of 2.7 Å (Egloff et al. 2004) and the other to 2.8 Å (Sutton et al. 2004). The studies consistently established that nsp9 is a single-strand RNA/DNA binding protein. The structure of SARS-CoV nsp9 has a central core comprised of seven β-strands. The core is flanked by a C-terminal α-helix and an N-terminal extension (Fig. 7.4a). X-ray crystallography, dynamic light scattering, analytical ultracentrifugation and GST pull-down experiments indicate that nsp9 forms dimers in solution (Campanacci et al. 2003; Imbert et al. 2008; Sutton et al. 2004). RNA binding by nsp9 has been suggested to involve the loops of the β-barrel domain while the C-terminal β-hairpin and helix, which are well conserved across coronaviruses, are probably involved in dimerization and interactions with other proteins. Database searches did not reveal structural homologs for nsp9 (Egloff et al. 2004). However, the short six-stranded β-barrel of nsp9 includes an open five-stranded barrel that is reminiscent of the five-stranded β-barrel structure of oligosaccharide/oligonucleotide-binding (OB)-fold proteins. About two-thirds of the proteins belonging to this protein superfamily are nucleic acid-binding proteins (Arcus 2002). As in OB-fold proteins, SARS-CoV nsp9 appears to bind nucleic acids by using a network of positively charged amino acids for binding the phosphate backbone of the substrate, whereas several exposed aromatic residues probably make additional stacking interactions with nucleobases (Fig. 7.4b). The nucleic acid-binding properties of nsp9 were subsequently confirmed by surface plasmon resonance, fluorescence quenching studies (Egloff et al. 2004) and electrophoretic mobility shift assays (Sutton et al. 2004). To date, there is no evidence to suggest that the nucleic acid-binding activity of nsp9 is sequence-specific.
Ribbon representation of SARS-CoV nsp9 (from Egloff et al. 2004; PDB 1QZ8). (a) One molecule of the dimer is yellow and the other is blue. (b) Electrostatic surface potential of nsp9 is colored according to electrostatic potential (blue, positive charge; red, negative charge). Images were generated using PYMOL
SARS-CoV nsp9 was shown to interact with nsp6, nsp7 and nsp8 (von Brunn et al. 2007). Nsp9/nsp8 interactions were confirmed by a number of methods, including analytical ultracentrifugation, yeast two-hybrid data, GST pull-down and co-immunoprecipitation experiments, suggesting quite stable interactions between the two proteins. Moreover, the structural disorder of the nsp8 N-terminal region in solution has been reported to decrease when nsp9 was added to a solution containing nsp8 (Sutton et al. 2004). On the basis of the nsp7–nsp8 hexadecameric structure (see Sect. 7.2.2), it was suggested that the most probable nsp9 binding site is formed by the N-terminal 50 residues of nsp8. This part of the structure is close to the entrance of the central pore and appears to be quite flexible, as indicated by the lack of electron density for these residues. Both the nsp9 structure and tryptophan fluorescence quenching data suggest that ssRNA is wrapped around the nsp9 dimer (Egloff et al. 2004). It has been speculated that nsp9 dimers bind to single-stranded nascent and template strands as they emerge from the channel of the nsp7–nsp8 complex at a time when stable secondary structures have not yet formed, thereby protecting ssRNAs from ribonucleolytic cleavage.
2.4 The nsp10 Protein
Nsp10 is well conserved among coronaviruses. In the polyprotein 1a, the protein is located upstream of nsp11, a short 13-residue peptide encoded by the 3′-terminal nucleotides of ORF1a. Nsp10 is thought to have an essential role in viral replication. It was shown that a MHV temperature-sensitive mutant carrying a nonsynonymous mutation in the nsp10 coding sequence has a defect in minus-strand RNA synthesis at the nonpermissive temperature (Sawicki et al. 2005; Siddell et al. 2001). Crystallization of nsp10 revealed monomers and homodimers (Joseph et al. 2006) whereas a complex dodecameric structure was observed when nsp10 was expressed and crystallized as a fusion with nsp11 (Su et al. 2006). The nsp10 monomer consists of a pair of antiparallel N-terminal helices stacked against an irregular β-sheet, a coil-rich C-terminus, and two zinc fingers (Fig. 7.5). Nsp10 represents a novel fold and is the first structural representative of this family of zinc finger proteins. The zinc finger motifs are strictly conserved in coronaviruses, supporting their essential role in viral replication.
Ribbon diagram of SARS-CoV nsp10 (from Joseph et al. 2006; PDB 2FYG). Residues coordinating the two Zn2+ ions are shown as sticks and pink balls, respectively. Atoms are colored as follows: green, carbon; blue, nitrogen; red, oxygen; orange, sulfur
It is currently unclear whether or not the nsp10 dodecameric structure seen in one of the structural studies is of biological relevance. The relevance has been questioned for several reasons. First, site-directed mutagenesis of the nsp10–nsp11 cleavage site in the MHV genome generated nonviable viruses, indicating that 3CLpro-mediated cleavage at this site is essential for viral replication (Deming et al. 2007). Second, the monomer structure has an intact second zinc finger which appears to stabilize the structure of the C-terminal tail of nsp10. By contrast, in the dodecamer structure, the second zinc finger lacks the C-proximal cysteine residue, resulting in local disorder at the nsp10 carboxyl terminus. Finally, substitutions of residues predicted to be crucial for the dodecamer formation did not cause a lethal phenotype in MHV (Donaldson et al. 2007).
Gel shift assays indicate that nsp10 binds single- and double-stranded RNA and DNA with low affinity and without obvious sequence specificity (Joseph et al. 2006). SARS-CoV nsp10 was shown to interact with nsp9 (Su et al. 2006). It also interacts strongly with nsp14 and nsp16 and, to a lesser extent, with nsp8 (Imbert et al. 2008). The precise role of nsp10 within the coronavirus replication/transcription complex remains to be identified.
3 The Endoribonuclease, nsp15
The uridylate-specific endoribonuclease (nsp15 or NendoU) is conserved in all members of the Nidovirales but no other virus, which makes the protein a major genetic marker of this group of viruses (Gorbalenya et al. 2006; Ivanov et al. 2004a). Distantly-related homologs of NendoU were also identified in some prokaryotes and eukaryotes, where they form a small protein family prototyped by XendoU, a Xenopus laevis endoribonuclease involved in the nucleolytic processing of intron-encoded box C/D U16 small nucleolar RNAs (Laneve et al. 2003; Snijder et al. 2003). Mainly on the basis of this sequence relationship, it has been speculated that NendoU may (also) have cellular substrates and it should be interesting to investigate whether NendoU is involved in producing small regulatory RNAs (similar to the small nucleolar RNAs produced by XendoU). Nsp15 activity is significantly stimulated by manganese ions and the enzyme generates 2′–3′ cyclic phosphate ends (Ivanov et al. 2004a). It is generally accepted that nsp15 functions as a homohexamer, even though the enzyme has some activity as a monomer (Guarino et al. 2005; Ivanov et al. 2004a; Joseph et al. 2007; Ricagno et al. 2006). NendoU cleaves downstream of uridylates, both in single and double-stranded RNA. The sequence context of the uridylate has been shown to affect cleavage efficiency and differential cleavage efficiencies have been reported for base-paired and nonbase-paired uridylates, respectively (Bhardwaj et al. 2006; Ivanov et al. 2004a).
Nsp15 crystal structures have been reported for SARS-CoV (Fig. 7.6) (Joseph et al. 2007; Ricagno et al. 2006; Bhardwaj et al. 2008) and MHV (Xu et al. 2006). Nsp15 exhibits a unique fold and assembles into a toric hexameric structure (Fig. 7.6b) with a central pore and six potentially active catalytic sites at the periphery. Unlike the situation in the nsp7–nsp8 complex, the diameter of the central pore seen in the nsp15 hexamer is too small to accommodate RNA. Furthermore, this part of the structure does not appear to interact with RNA (Bhardwaj et al. 2006; Ricagno et al. 2006). Each protomer contains nine α-helices and 21 β-strands (Fig. 7.6a). Alanine substitutions of highly conserved residues demonstrated that the C-terminal domain contains the active site. There are striking similarities between the active site residues of nsp15 and RNase A, suggesting that the enzymes use common catalytic mechanisms, although they are not related in their tertiary structures (Ricagno et al. 2006). The general acid–base mechanism used by coronavirus NendoUs probably involves His-234, His-249, and Lys-289 (SARS-CoV nsp15 numbering). Additional conserved residues likely involved in substrate specificity have been identified by X-ray crytallography and site-directed mutagenesis (Bhardwaj et al. 2008; Ricagno et al. 2006).
Structure of SARS-CoV nsp15 endoribonuclease (from Ricagno et al. 2006; PDB 2H85). (a) Cartoon representation of the nsp15 monomer. The structure consists of three domains: N-terminal domain (α1–α2), central domain (α3–β8), and C-terminal domain (α6–β14). Secondary structures are colored as follows: red, α-helices; yellow, β-sheets; and green, loops. (b) A view of the nsp15 hexamer. In the trimer shown in the foreground, the subdomains of one of the monomers are colored as follows: green, N-terminal domain; yellow, central domain; and red, C-terminal domain. The other two molecules are shown in blue and magenta
The overall architecture of coronavirus NendoUs suggests that the protein might be tethered to other partners connecting this hexameric assembly to the viral replication complex and, potentially, cellular structures. Consistent with this idea, NendoU has been implicated in coronavirus and arterivirus RNA synthesis and/or the production of virus progeny (Ivanov et al. 2004a; Kang et al. 2007; Posthuma et al. 2006) and interactions between SARS-CoV nsp15 and nsp3, nsp8, nsp9, and nsp12 have been reported (Imbert et al. 2008). Although endonucleases are ubiquitous enzymes that are involved in many aspects of RNA metabolism, they are extremely rare in the RNA virus world, with only very few exceptions (e.g., in orthomyxo-, pesti-, and retroviruses). Although the precise role of the enzyme in the viral life cycle and its biologically relevant substrates remain to be identified, it seems reasonable to think that NendoU acts as part of a larger protein complex and that nucleolytic activity and substrate specificity are tightly regulated by additional factors to minimize uncontrolled nucleolytic cleavage of viral and cellular RNAs.
4 Conclusion
The SARS outbreak in 2003 sparked significant new interest in the molecular biology and biochemistry of coronavirus replication. A multitude of structural and functional studies into SARS-CoV replicative enzymes have been published over the past few years, giving a major boost to coronavirus and, more generally, nidovirus research. Interestingly, in several instances, crystal structures of coronavirus proteins have uncovered new biochemical and enzymatic activities, thus opening up new vistas for future studies and stimulating exciting biochemical and genetic studies into the biological functions of specific coronavirus nonstructural proteins. Also, the SARS-CoV structural work has been extremely rewarding for crystallographers interested in novel folds and original enzymes. There are still major challenges ahead, mainly with respect to ORF1b-encoded proteins. For example, there is still no crystal structure available for the special type of primer-dependent RNA polymerases encoded by coronaviruses. Also studies of the zinc-binding domain-containing superfamily 1 helicase (Ivanov et al. 2004b) and 3′–5′ exonuclease (Minskaia et al. 2006) domains residing in nsp13 and nsp14, respectively, and the functionally interesting nsp16 2′-O-methyltransferase (Decroly et al. 2008) promise to reveal exciting new structural and functional insight. Undoubtedly, these upcoming structures will greatly stimulate coronavirus (nidovirus) research and perhaps even reveal novel and truly unique RNA processing pathways involved in the replication of these viruses.
References
Arcus V (2002) OB-fold domains: a snapshot of the evolution of sequence, structure and function. Curr Opin Struct Biol 12:794–801
Barretto N, Jukneliene D, Ratia K, Chen Z, Mesecar AD, Baker SC (2005) The papain-like protease of severe acute respiratory syndrome coronavirus has deubiquitinating activity. J Virol 79:15189–15198
Bhardwaj K, Sun J, Holzenburg A, Guarino LA, Kao CC (2006) RNA recognition and cleavage by the SARS coronavirus endoribonuclease. J Mol Biol 361:243–256
Bhardwaj K, Palaninathan S, Alcantara JM, Yi LL, Guarino L, Sacchettini JC, Kao CC (2008) Structural and functional analyses of the severe acute respiratory syndrome coronavirus endoribonuclease Nsp15. J Biol Chem 283:3655–3664
Bost AG, Carnahan RH, Lu XT, Denison MR (2000) Four proteins processed from the replicase gene polyprotein of mouse hepatitis virus colocalize in the cell periphery and adjacent to sites of virion assembly. J Virol 74:3379–3387
Brockway SM, Lu XT, Peters TR, Dermody TS, Denison MR (2004) Intracellular localization and protein interactions of the gene 1 protein p28 during mouse hepatitis virus replication. J Virol 78:11551–11562
Campanacci V, Egloff MP, Longhi S, Ferron F, Rancurel C, Salomoni A, Durousseau C, Tocque F, Bremond N, Dobbe JC, Snijder EJ, Canard B, Cambillau C (2003) Structural genomics of the SARS coronavirus: cloning, expression, crystallization and preliminary crystallographic study of the Nsp9 protein. Acta Crystallogr D Biol Crystallogr 59:1628–1631
Decroly E, Imbert I, Coutard B, Bouvet M, Selisko B, Alvarez K, Gorbalenya AE, Snijder EJ, Canard B (2008) Coronavirus nonstructural protein 16 is a cap-0 binding enzyme possessing (nucleoside-2'O)-methyltransferase activity. J Virol 82(16):8071–8084
Deming DJ, Graham RL, Denison MR, Baric RS (2007) Processing of open reading frame 1a replicase proteins nsp7 to nsp10 in murine hepatitis virus strain A59 replication. J Virol 81:10280–10291
Donaldson EF, Sims AC, Graham RL, Denison MR, Baric RS (2007) Murine Hepatitis Virus replicase protein nsp10 is a critical regulator of viral RNA synthesis. J Virol 81(12):6356–6368
Egloff MP, Ferron F, Campanacci V, Longhi S, Rancurel C, Dutartre H, Snijder EJ, Gorbalenya AE, Cambillau C, Canard B (2004) The severe acute respiratory syndrome-coronavirus replicative protein nsp9 is a single-stranded RNA-binding subunit unique in the RNA virus world. Proc Natl Acad Sci USA 101:3792–3796
Egloff MP, Malet H, Putics A, Heinonen M, Dutartre H, Frangeul A, Gruez A, Campanacci V, Cambillau C, Ziebuhr J, Ahola T, Canard B (2006) Structural and functional basis for ADP-ribose and poly(ADP-ribose) binding by viral macro domains. J Virol 80:8493–8502
Gorbalenya AE, Koonin EV, Lai MM (1991) Putative papain-related thiol proteases of positive-strand RNA viruses. Identification of rubi- and aphthovirus proteases and delineation of a novel conserved domain associated with proteases of rubi-, alpha- and coronaviruses. FEBS Lett 288:201–205
Gorbalenya AE, Pringle FM, Zeddam JL, Luke BT, Cameron CE, Kalmakoff J, Hanzlik TN, Gordon KH, Ward VK (2002) The palm subdomain-based active site is internally permuted in viral RNA-dependent RNA polymerases of an ancient lineage. J Mol Biol 324:47–62
Gorbalenya AE, Enjuanes L, Ziebuhr J, Snijder EJ (2006) Nidovirales: evolving the largest RNA virus genome. Virus Res 117:17–37
Guarino LA, Bhardwaj K, Dong W, Sun J, Holzenburg A, Kao C (2005) Mutational analysis of the SARS virus Nsp15 endoribonuclease: identification of residues affecting hexamer formation. J Mol Biol 353:1106–1117
Harcourt BH, Jukneliene D, Kanjanahaluethai A, Bechill J, Severson KM, Smith CM, Rota PA, Baker SC (2004) Identification of severe acute respiratory syndrome coronavirus replicase products and characterization of papain-like protease activity. J Virol 78:13600–13612
Imbert I, Guillemot JC, Bourhis JM, Bussetta C, Coutard B, Egloff MP, Ferron F, Gorbalenya AE, Canard B (2006) A second, non-canonical RNA-dependent RNA polymerase in SARS coronavirus. Embo J 25:4933–4942
Imbert I, Snijder EJ, Dimitrova M, Guillemot JC, Lecine P, Canard B (2008) The SARS-Coronavirus PLnc domain of nsp3 as a replication/transcription scaffolding protein. Virus Res 133:136–148
Ivanov KA, Hertzig T, Rozanov M, Bayer S, Thiel V, Gorbalenya AE, Ziebuhr J (2004a) Major genetic marker of nidoviruses encodes a replicative endoribonuclease. Proc Natl Acad Sci USA 101:12694–12699
Ivanov KA, Thiel V, Dobbe JC, van der Meer Y, Snijder EJ, Ziebuhr J (2004b) Multiple enzymatic activities associated with severe acute respiratory syndrome coronavirus helicase. J Virol 78:5619–5632
Joseph JS, Saikatendu KS, Subramanian V, Neuman BW, Brooun A, Griffith M, Moy K, Yadav MK, Velasquez J, Buchmeier MJ, Stevens RC, Kuhn P (2006) Crystal structure of nonstructural protein 10 from the severe acute respiratory syndrome coronavirus reveals a novel fold with two zinc-binding motifs. J Virol 80:7894–7901
Joseph JS, Saikatendu KS, Subramanian V, Neuman BW, Buchmeier MJ, Stevens RC, Kuhn P (2007) Crystal structure of a monomeric form of severe acute respiratory syndrome coronavirus endonuclease nsp15 suggests a role for hexamerization as an allosteric switch. J Virol 81:6700–6708
Kang H, Bhardwaj K, Li Y, Palaninathan S, Sacchettini J, Guarino L, Leibowitz JL, Kao CC (2007) Biochemical and genetic analyses of murine hepatitis virus Nsp15 endoribonuclease. J Virol 81:13587–13597
Kanjanahaluethai A, Chen Z, Jukneliene D, Baker SC (2007) Membrane topology of murine coronavirus replicase nonstructural protein 3. Virology 361:391–401
Laneve P, Altieri F, Fiori ME, Scaloni A, Bozzoni I, Caffarelli E (2003) Purification, cloning, and characterization of XendoU, a novel endoribonuclease involved in processing of intron-encoded small nucleolar RNAs in Xenopus laevis. J Biol Chem 278:13026–13032
Lindner HA, Fotouhi-Ardakani N, Lytvyn V, Lachance P, Sulea T, Menard R (2005) The papain-like protease from the severe acute respiratory syndrome coronavirus is a deubiquitinating enzyme. J Virol 79:15199–15208
Martzen MR, McCraith SM, Spinelli SL, Torres FM, Fields S, Grayhack EJ, Phizicky EM (1999) A biochemical genomics approach for identifying genes by the activity of their products. Science 286:1153–1155
Minskaia E, Hertzig T, Gorbalenya AE, Campanacci V, Cambillau C, Canard B, Ziebuhr J (2006) Discovery of an RNA virus 3′-> 5′ exoribonuclease that is critically involved in coronavirus RNA synthesis. Proc Natl Acad Sci USA 103:5108–5113
Ng LF, Xu HY, Liu DX (2001) Further identification and characterization of products processed from the coronavirus avian infectious bronchitis virus (IBV) 1a polyprotein by the 3C-like proteinase. Adv Exp Med Biol 494:291–298
Pasternak AO, Spaan WJ, Snijder EJ (2006) Nidovirus transcription: how to make sense...? J Gen Virol 87:1403–1421
Pehrson JR, Fried VA (1992) MacroH2A, a core histone containing a large nonhistone region. Science 257:1398–1400
Peti W, Johnson MA, Herrmann T, Neuman BW, Buchmeier MJ, Nelson M, Joseph J, Page R, Stevens RC, Kuhn P, Wuthrich K (2005) Structural genomics of the severe acute respiratory syndrome coronavirus: nuclear magnetic resonance structure of the protein nsP7. J Virol 79:12905–12913
Posthuma CC, Nedialkova DD, Zevenhoven-Dobbe JC, Blokhuis JH, Gorbalenya AE, Snijder EJ (2006) Site-directed mutagenesis of the Nidovirus replicative endoribonuclease NendoU exerts pleiotropic effects on the arterivirus life cycle. J Virol 80:1653–1661
Putics A, Filipowicz W, Hall J, Gorbalenya AE, Ziebuhr J (2005) ADP-ribose-1″-monophosphatase: a conserved coronavirus enzyme that is dispensable for viral replication in tissue culture. J Virol 79:12721–12731
Putics A, Gorbalenya AE, Ziebuhr J (2006) Identification of protease and ADP-ribose 1″-monophosphatase activities associated with transmissible gastroenteritis virus non-structural protein 3. J Gen Virol 87:651–656
Ratia K, Saikatendu KS, Santarsiero BD, Barretto N, Baker SC, Stevens RC, Mesecar AD (2006) Severe acute respiratory syndrome coronavirus papain-like protease: structure of a viral deubiquitinating enzyme. Proc Natl Acad Sci USA 103:5717–5722
Ricagno S, Egloff MP, Ulferts R, Coutard B, Nurizzo D, Campanacci V, Cambillau C, Ziebuhr J, Canard B (2006) Crystal structure and mechanistic determinants of SARS coronavirus nonstructural protein 15 define an endoribonuclease family. Proc Natl Acad Sci USA 103:11892–11897
Saikatendu KS, Joseph JS, Subramanian V, Clayton T, Griffith M, Moy K, Velasquez J, Neuman BW, Buchmeier MJ, Stevens RC, Kuhn P (2005) Structural basis of severe acute respiratory syndrome coronavirus ADP-ribose-1″-phosphate dephosphorylation by a conserved domain of nsP3. Structure 13:1665–1675
Sawicki SG, Sawicki DL, Younker D, Meyer Y, Thiel V, Stokes H, Siddell SG (2005) Functional and genetic analysis of coronavirus replicase-transcriptase proteins. PLoS Pathog 1:e39
Siddell S, Sawicki D, Meyer Y, Thiel V, Sawicki S (2001) Identification of the mutations responsible for the phenotype of three MHV RNA-negative ts mutants. Adv Exp Med Biol 494:453–458
Snijder EJ, Bredenbeek PJ, Dobbe JC, Thiel V, Ziebuhr J, Poon LL, Guan Y, Rozanov M, Spaan WJ, Gorbalenya AE (2003) Unique and conserved features of genome and proteome of SARS-coronavirus, an early split-off from the coronavirus group 2 lineage. J Mol Biol 331:991–1004
Su D, Lou Z, Sun F, Zhai Y, Yang H, Zhang R, Joachimiak A, Zhang XC, Bartlam M, Rao Z (2006) Dodecamer structure of severe acute respiratory syndrome coronavirus nonstructural protein nsp10. J Virol 80:7902–7908
Sutton G, Fry E, Carter L, Sainsbury S, Walter T, Nettleship J, Berrow N, Owens R, Gilbert R, Davidson A, Siddell S, Poon LL, Diprose J, Alderton D, Walsh M, Grimes JM, Stuart DI (2004) The nsp9 replicase protein of SARS-coronavirus, structure and functional insights. Structure 12:341–353
Thiel V, Ivanov KA, Putics A, Hertzig T, Schelle B, Bayer S, Weissbrich B, Snijder EJ, Rabenau H, Doerr HW, Gorbalenya AE, Ziebuhr J (2003) Mechanisms and enzymes involved in SARS coronavirus genome expression. J Gen Virol 84:2305–2315
von Brunn A, Teepe C, Simpson JC, Pepperkok R, Friedel CC, Zimmer R, Roberts R, Baric R, Haas J (2007) Analysis of Intraviral Protein−Protein Interactions of the SARS Coronavirus ORFeome. PLoS ONE 2:e459
Xu X, Zhai Y, Sun F, Lou Z, Su D, Xu Y, Zhang R, Joachimiak A, Zhang XC, Bartlam M, Rao Z (2006) New antiviral target revealed by the hexameric structure of mouse hepatitis virus nonstructural protein nsp15. J Virol 80:7909–7917
Zhai Y, Sun F, Li X, Pang H, Xu X, Bartlam M, Rao Z (2005) Insights into SARS-CoV transcription and replication from the structure of the nsp7-nsp8 hexadecamer. Nat Struct Mol Biol 12:980–986
Ziebuhr J, Thiel V, Gorbalenya AE (2001) The autocatalytic release of a putative RNA virus transcription factor from its polyprotein precursor involves two paralogous papain-like proteases that cleave the same peptide bond. J Biol Chem 276:33220–33232
Zust R, Miller TB, Goebel SJ, Thiel V, Masters PS (2008) Genetic interactions between an essential 3′ cis-acting RNA pseudoknot, replicase gene products, and the extreme 3' end of the mouse coronavirus genome. J Virol 82:1214–1228
Acknowledgment
The work of I.I. and B.C. was supported by the Structural Proteomics in Europe (SPINE) project of the European Union 5th Framework Research Program (Grant QLRT-2001-00988) and subsequently by the Euro-Asian SARS-DTV Network (SP22-CT-2004-511064) from the European Commission Specific Research and Technological Development Program “Integrating and Strengthening the European Research Area” and by Interdisciplinary CNRS 2007 Program “Infectious Emerging Diseases.” The work of R.U. and J.Z. has been supported by the Sino-German Centre for Research Promotion, the German Research Council and the Queen’s University of Belfast.
Author information
Authors and Affiliations
Corresponding author
Editor information
Editors and Affiliations
Rights and permissions
Copyright information
© 2010 Springer-Verlag Berlin Heidelberg
About this chapter
Cite this chapter
Imbert, I., Ulferts, R., Ziebuhr, J., Canard, B. (2010). SARS Coronavirus Replicative Enzymes: Structures and Mechanisms. In: Lal, S. (eds) Molecular Biology of the SARS-Coronavirus. Springer, Berlin, Heidelberg. https://doi.org/10.1007/978-3-642-03683-5_7
Download citation
DOI: https://doi.org/10.1007/978-3-642-03683-5_7
Published:
Publisher Name: Springer, Berlin, Heidelberg
Print ISBN: 978-3-642-03682-8
Online ISBN: 978-3-642-03683-5
eBook Packages: Biomedical and Life SciencesBiomedical and Life Sciences (R0)