Abstract
The diatoms are unicellular algae of great interest in paleontology, aquatic ecology, and forensic medicine, among others. Currently, there are more than 100 000 known species distributed in aquatic ecosystems. For that reason, there is a big interest in the automatic classification of diatom images, however, the preliminary process applied to isolate the diatom from the background is a complex task. In this paper, we propose a segmentation method and an object-extraction procedure to extract the diatom from the background. First, we binarize the image by searching the optimal threshold in the histogram based on its cumulative distribution function. Then we eliminate, under some spatial criteria, all regions other than those that could be part of the diatom. Afterwards, we construct the convex hull of all remaining components. Finally, from this first polygonal approximation, we construct the diatom contour by successive refinements of the convex hull shape.
You have full access to this open access chapter, Download conference paper PDF
Similar content being viewed by others
Keywords
1 Introduction
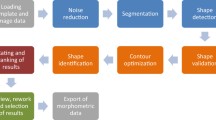
The diatoms shell, or the frustule, consists of two embedded silica valves. The identification of a diatom, made by a specialist under a light binocular microscope, is based on the recognition of specific patterns, like shape parameters and internal features. The principal problem working with a diatom image is the low contrast followed by light reflections and blur (Fig. 1 (a)). In some cases there are also isolated particles around the diatom or other fragmented diatoms hiding partially the specimen to identify (Fig. 1 (b) and (c)). In image processing, threshold and object extraction are very important tasks as well as some of the most complex. Many threshold techniques have been developed concerning diatoms, but none of the methods are capable of getting high performance in large conditions. Some works on diatoms classification [1,2,3,4] have focused on feature extraction and classification but none in the preliminary process of image segmentation. In this paper, we propose a segmentation method and an object-detection procedure as preliminary processes in automatic diatoms classification. In Sect. 2, we present a segmentation method based on the unimodal binarization when there is a dominant heap in the middle of the grey level histogram. In Sect. 3, we propose a method to detect parts of the image not belonging to the diatom. In Sect. 4, we present the steps to reconstruct the diatom.
2 Segmentation
The segmentation of the diatom images is an important step before the extraction of the geometrical features we need for an accurate classification of diatom species. For instance, we have to characterize the shape contour of the diatom and identify internal features that characterize the species. Several segmentation methods are based on one or more specific assumptions. Many of these methods aim to binarize the image and assume that there are only two independent classes of pixel intensities. We have applied three standard methods and adapted the Rosin’s one. We tested first the method of Balanced Histogram Threshold (BHT) [5], and the Otsu method [6] (see Fig. 2 (b) and (c)). Furthermore, we tested the K-Means method and we obtained similar results as the Otsu one. The method developed by Rosin [7] consists in splitting pixels in two classes from a unimodal distribution in the histogram of grey levels. The method assumes that there exists only one dominant heap in the histogram. If we suppose that the heap is at the left in the distribution, a straight line L is drawn from the highest position on the heap to the last empty bin on the right of the histogram. If \(H_i\) is the value of the level \({ t_i}\) of the histogram, then the threshold corresponds to the value \({ t_i}\) where the distance between the secant line L and the point \((t_i, H_i)\) is maximum (see Fig. 3).
Rosin method to determine the abrupt change on the histogram [7].
In this paper, we make assumptions based on features shared by diatoms images (Fig. 1). First, we observe that the background is quasi-uniform and any isolated region is small compared to the diatom. Second, we do not consider images with diatoms partially covered by another diatom or cut out by the image frame. Finally, we suppose that there is only one dominant heap in the histogram corresponding to the background. Thus, our segmentation approach consists in finding two optimal thresholds at both sides of the heap. The intensities between those thresholds belong to the background and the others, to the diatom. Our segmentation method is, thus, based on the Rosin methodology for a unimodal distribution of grey levels, but we applied this approach with two principal differences. First, since the heap is in the middle of the histogram, we determined two thresholds; this is why we do not use the Rosin method as it is. Second, we apply the methodology to the cumulative distribution function (CDF), which is near a sigmoid function in this case. Approximating the CDF by a sigmoid function allows us to locate the two inflection points around the thresholds.
Finally, since the CDF is relatively smooth, we interpolate its curve by a sigmoid function (SCDF) on which the Rosin method can be adapted to our context. Our objective being the determination of two optimal values of grey level as thresholds, we draw two straight lines in both sides of the SCDF curve. A first line \(L_l\) starts at the greatest abscissa of the first consecutive zero bins and it ends at the first tangent coordinates at the right on the SCDF. A second line \(L_r\) starts at the last point on the SCDF curve and it ends at the point where the line is still tangent to the curve and below it. Similarly to the Rosin procedure, the two thresholds \(t_l\) and \(t_r\) are the abscissas such that the perpendicular distances between the straight lines and the function are maximum as in the Fig. 4 (b). Finally, we binarize the image using the rule that if x is a pixel with intensity \(t_x\) then if \(t_{l} \le t_x \le t_{r}\) we set the intensity equal to 0, otherwise the intensity is set at 255. Afterwards, in order to improve the results of the segmentation, we apply successively the morphological operators closing and opening based on a circular structural element of size three and five respectively. In Fig. 4 (b), we see the resulting thresholds based on the histogram corresponding to the image in Fig. 2 (a).
3 Detection of Diatom Fragments
A binary image of a diatom contains isolated white fragments with a black background (Fig. 4 (a)). In such an image, we have to identify the subset of fragments that belong to the diatom and eliminate the others, caused by small particles, light reflects and shadows. For that, we first label all connected components in the image and we identify the biggest one.Then, we normalize the size of each component by the area of the convex hull of the biggest one. After, we calculate the minimal Euclidean distance between the biggest component and the others. We reduce the execution time by ignoring components inside the convex hull of another component. Each distance is then normalized by the half of the length of the image diagonal. We define \(s_1, s_{2},\dots , s_i,\dots , s_n\) as the area of the n components. If \(s_1\) is the biggest area then \(s_1=1\). We also define \(d_1, d_{2},\dots , d_i,\dots d_{n}\) as the distances between each component and the biggest one, then \(d_1\) must be zero. At this step, we calculate the force of attraction between a component and the biggest component, in order to eliminate particles outside the diatom. This force is given by,
\(F_i = \displaystyle G\frac{s_1 s_i}{d_i^2}=\displaystyle \frac{s_i}{d_i^2},2\le i \le n,\)
where G is the gravitational constant. In our application we set \(G=1\). Empirically, we have rejected particles with an force of attraction lower than \(\sigma = 40\). The other particles are considered to be part of the principal diatom \(s_1\) (Fig. 4).
4 Diatom Reconstruction
Ideally, the latest procedure would give us a single connected component on a black background. However, in many cases we obtain an object with several components close to each other, but keeping the shape of a diatom. From the resulting image, we will construct a polygonal contour of the combined components in order to obtain an identifiable diatom. In many works, contour is detected with an active contour. Nevertheless, the snake method does not work well in our case because of the dispersed components. So, first, we choose to initialize the construction of the contour with the convex hull [8].
After this initialization, we refine the contour locally and step by step. Such a refinement procedure is necessary to precise an adequate shape of diatom (Fig. 5). For that purpose, we identify a reference set \(\varGamma \) of clockwise ordered contour pixels. The points in \(\varGamma \) are the first and last ones in the intersection of a vertical or a horizontal line intersecting the components. The convex hull of the diatom components is now defined by an ordered set of border points \(C = \{p_{1}, p_{2}, \cdots , p_{n}=p_1\}\subset \varGamma \) and each segment \(\ell _{i}\) in the hull is determined by the pixels \(p_{i}\) and \(p_{i + 1}\) for \(1\le i \le n-1\). The refinement of the contour will be done only on a subset \(\mathcal S\) of hull segments and such that \(\ell _{i}\) in this subset has a length outside the whiskers resulting from a box plot on all segments lengths. For each segment \(\ell _{k}\) in \(\mathcal {S}\), we identify a subset \(\varGamma _k \subset \varGamma \) of border points that could be selected to refine locally the segment. These points are those in the same side of the two half planes defined by two directions originated from the extremities and corresponding to the mean orientations of the contour at each side, each one is calculated with the half of the contour pixels (Fig. 6). Then, for successive values \(m=2,3,\dots \), we split the indices of the points in \(\varGamma _k\), between \(p_k\) and \(p_{k+1}\), in m subsets of equal cardinality. The extremities of each subset of indices, the last index of a subset being the first one of the next subset, define new subsegments added to the polygonal approximation of the contour. At step m, we verify if all the lengths of these new subsegments is lower or equal to the mean of the initial contour; if not, we increase the value of m until the last criteria is satisfied. The refinement procedure is illustrated in Fig. 6. After the refinement of all the segments in \(\mathcal {S}\), the resulting polygonal is considered as the contour of the diatom.
5 Experimental Results
We tested our approach on 292 images with either quasi smooth background or isolated particles. For 11.64 % of samples the segmentation method failed. This is generally caused by a low contrast and light reflectance due to the capture of images. For the remaining 88.36 %, we obtained good results, 66.43 % of samples have a single significant component with small particles around. These particles were eliminated by our gravitation criteria when they were not both big and close to the principal component, otherwise the test is considered as a fail. For the other 21.91 % we obtained several components keeping the shape of the diatom. For these cases, the resulting convex hull joined together in a single component. In Fig. 7 we observe the original images with their segmented representations and the extracted diatom. For sample 1, we observe that the final contour keeps a part of the background. This loss of accuracy is caused by the shadows on the border of the diatom but it will not affect the classification process with respect to the shape parameters issued from the contour [1]. For most of the samples tested from our database, the segmentation and the extraction have produced good results as shown for the samples 2, 3 and 4. We can see that the brightness, the contrast and the general quality of the captured samples are important factors in the result of the segmentation process.
6 Conclusion
The aim of this work was the extraction of a diatom from the background after the image acquisition under a binocular microscope. This extraction is a preliminary and necessary step in an automatic classification process. In [1], the authors showed that a classification, by grouping species according to their similar contour shapes, is promising. However, the contrast enhancement and brightness equilibrium to improve the quality of the images needed further work. So the proposed extraction of interior features will facilitate the classification of different species.
References
Siddour, A., Nouboud, F., Mammass, D., Chalifour, A., Campeau, S.: Classification des diatomées par les descripteurs de Fourier du contour. Spécial MCSEAI 2006, e-TI, Numéro 4 / Issue 4 (2007)
Jalba, A.C., Wilkinson, M.H.F., Roerdink, J.B.T.M.: Automatic segmentation of diatom images for classification. Microsc. Res. Tech. 65, 72–85 (2004)
Álvarez, T., Martín, Y., Pérez, S., Santos, F., Tadeo, F., González, S., Arribas, J.L., Vega, P.: Classification of microorganisms using image processing techniques. In: Proceedings of International Conference on Image Processing (Greece), pp. 329–332 (2001)
Ciobanu, A., Shahbazkia, H., du Buf, H.: Contour profiling by dynamic ellipse fitting. In: Proceedings of 15th International Conference on Pattern Recognition, pp. 750–753 (2000)
Anjos, A., Shahbazkia, H.: Bi-level image thresholding - a fast method. In: Biosignals, pp. 70–76 (2008)
Otsu, N.: A threshold selection method from gray-level histograms. IEEE Trans. Syst. Man Cybern. 9, 62–66 (1979)
Rosin, P.L.: Unimodal thresholding. J. Pattern Recognit. Soc. 34, 2083–2096 (2001)
Barber, C.B., Dobkin, D.P., Huhdanpaa, H.: The quickhull algorithm for convex hulls. ACM Trans. Math. Softw. 22, 469–483 (1996)
Author information
Authors and Affiliations
Corresponding author
Editor information
Editors and Affiliations
Rights and permissions
Copyright information
© 2018 Springer International Publishing AG, part of Springer Nature
About this paper
Cite this paper
Gutiérrez Lira, E., Nouboud, F., Chalifour, A., Voisin, Y. (2018). Image Segmentation and Object Extraction for Automatic Diatoms Classification. In: Mansouri, A., El Moataz, A., Nouboud, F., Mammass, D. (eds) Image and Signal Processing. ICISP 2018. Lecture Notes in Computer Science(), vol 10884. Springer, Cham. https://doi.org/10.1007/978-3-319-94211-7_7
Download citation
DOI: https://doi.org/10.1007/978-3-319-94211-7_7
Published:
Publisher Name: Springer, Cham
Print ISBN: 978-3-319-94210-0
Online ISBN: 978-3-319-94211-7
eBook Packages: Computer ScienceComputer Science (R0)