Abstract
Clinical information systems store a large amount of data in medical databases. In the use of medical dataset for diagnosis, the patient’s information is selectively collected and interpreted based on previous knowledge for detecting the existence of disorders. Feature selection is important and necessary data pre-processing step in medical data classification process. In this work, we propose a wrapper method for feature subset selection based on a binary version of the Firefly Algorithm combined with the SVM classifier, which tries to reduce the initial size of medical data and to select a set of relevant features for enhance the classification accuracy of SVM. The proposed method is evaluated on some medical dataset and compared with some well-known classifiers. The computational experiments show that the proposed method with optimized SVM parameters provides competitive results and finds high quality solutions.
You have full access to this open access chapter, Download conference paper PDF
Similar content being viewed by others
Keywords
- Medical data classification
- Machine learning
- Feature selection
- Binary firefly algorithm
- Support vector machine (SVM)
- Cross-validation
1 Introduction
Clinical information systems store a large amount of information in medical databases. So, the manual classification of this information is becoming more and more difficult. Therefore, there is an increasing interest in developing automated evaluation methods to follow up the diseases. Classification is one of the techniques of data mining which involves extracting a general rule or classification procedure from a set of learning examples. Medical data classification refers to learning classification models from medical datasets and aims to improve the quality of health care [3].
As medical datasets are generally characterized as having high dimensionality, and many of the feature attributes in a typical medical dataset are collected for reasons other than data classification. Some of the features are redundant while others are irrelevant adding more noise to the dataset, although in medical diagnosis, it is desirable to select the clinical tests that have the least cost and risk and that are significantly important in determining the class of the disease [1].
Feature selection is important and necessary data pre-processing steps to increase the quality of the feature space. It aims to select a small subset of important (relevant) features from the original full feature set. It can potentially improve the performance of a learning algorithm significantly in terms of the accuracy; increase the learning speed, and simplifying the interpretation of the learnt models [2, 12]. Feature selection is used in different tasks of learning or data mining, in the fields of image processing, pattern recognition, data analysis in bioinformatics, categorization of texts, etc.
The methods used to evaluate a feature subset in the selection algorithms can be classified into three main approaches: filter methods, wrapper methods and embedded methods. Filter methods perform the evaluation independently of any classification algorithm; they are based on data and attributes [2]. Wrapper methods use the learning algorithm as an evaluation function. It therefore defines the relevance of the attributes through a prediction of the performance of the final system. Embedded methods combine the exploration process with a learning algorithm. The difference with wrapper methods is that the classifier not only serves to evaluate a candidate sub-set, but also to guide the selection mechanism.
Wrapper approaches conducts a search in the space of candidate subsets of features, and the quality of a candidate subset is evaluated by the performance of the classification algorithm trained on this subset [15]. Several wrapper methods have been proposed for feature selection, among them, the stochastic local search methods and the population based optimization metaheuristic methods, like the Genetic Algorithms (GA), the Memetic Algorithm (MA), Particle Swarm Optimization (PSO), and the Harmony Search Algorithm (HAS) [16].
In this work, we apply a wrapper method based on a binary version of Firefly Algorithm to the feature selection problem in medical data classification, in order to extract an ideally minimal subset of features with strong discriminative power. The proposed approach uses the SVM classifier for evaluating a feature subset.
This paper is organized as follows: first, we briefly outline the main idea of support vector machine methods (SVM) and the Binary Firefly Algorithm in Sect. 2. In Sect. 3, we describe the proposed approach for feature selection and classification of medical data. The experimental results are presented and discussed in Sect. 4. Finally, we conclude this study and discuss possible future work in Sect. 5.
2 Background
2.1 Support Vector Machines
Support Vector Machines (SVM) are a class of supervised learning algorithms introduced by Vladimir Vapnik [4]. The main principle of SVM is the construction of a function f called decision function that for an input vector x matches a value y, y = f (x), where x is the example to classify and y is the class which corresponds to the example input. SVM are originally defined for binary classification problems, and their extension to nonlinear problems is offered introducing the kernel functions. SVM are widely used in statistical learning and has proved effective in many application areas such as image processing, speech processing, bioinformatics, natural language processing, and even data sets of very large dimensions [13].
SVM classifiers are based on two key ideas: the notion of maximum margin and the concept of kernel function. The first key idea is the concept of maximum margin. We seek the hyperplane that separates the positive examples of negative examples, ensuring that the distance between the separation boundary and the nearest samples (margin) is maximal, they are called support vector. And as it seeks to maximize the margin, we will talk about wide margin separators [14].
The second key idea in SVM is the concept of kernel function. This is transforming the data space entries in a space of larger dimension called feature space in which it is likely that there is a dividing line, in order to deal with cases where the data are not linearly separable. Some examples of kernel functions are:
– Linear kernel: \( K\left( {x_{i} ,\,x_{j} } \right) = x_{i} \,.\,x_{j} \)
– Polynomial kernel: \( K\left( {x_{i} ,\,x_{j} } \right) = \left( {\gamma x_{i} \,.\,x_{j} \, + \,r} \right)^{d} ,\,\gamma > 0. \)
– RBF kernel: \( K\left( {x_{i} ,x_{j} } \right) = e^{{ - \frac{{\left| {x_{i} \, - \,x_{j} } \right|^{2} }}{{2\gamma^{2} }}}} ,\,\gamma > 0. \)
– Sigmoid kernel: \( K\left( {x_{i} ,x_{j} } \right) = tanh\left( {\gamma x_{i} \,.\,x_{j} + r} \right). \)
Where \( \gamma \), r and d are kernel parameters. In this study, we utilized the LIBSVM toolset and chose Radial Basis Function (RBF) as the kernel function, and its C and γ parameters are optimized using an iterative search method. Previous studies show that these two parameters play an important role on the success of SVMs [8].
2.2 Binary Firefly Algorithm
The Firefly algorithm is a recent bio-inspired metaheuristic developed by Xin She Yang in 2008 and it has become an important tool for solving the hardest optimization problems in almost all areas of optimization [6]. The algorithm is based on the principle of attraction between fireflies and simulates the behavior of a swarm of fireflies in nature, which gives it many similarities with other meta-heuristics based on the collective intelligence, such as the PSO (Particle Swarm Optimization) algorithm or the bee colony optimization algorithm. He uses the following three idealized rules:
-
All fireflies are unisex, meaning that one firefly is attracted by another, regardless of sex.
-
The attractiveness and brightness are proportional, so that for two flashing fireflies, the less bright will move towards the brighter. Attractiveness and brightness decrease with increasing distance. If there is not one firefly brighter than the other, they will place themselves randomly.
-
The brightness of a firefly is determined by the point of view of the objective function to optimize. For a maximization problem, the luminosity is simply proportional to the value of the objective function.
Since the attractiveness is proportional to the luminosity of the adjacent fireflies, then the variation in the attractiveness β with the distance r is defined by:
Where β 0 is the attractiveness at r = 0 and γ is the absorption coefficient.
The distance r ij between 2 fireflies is determined by the formula (2).
Where \( X_{i}^{k} \) is the kth component of the spatial coordinate of the ith firefly and d is the number of dimensions.
The movement of a firefly \( {\text{X}}_{\text{i}} \) to another firefly \( {\text{X}}_{\text{j}} \) more attractive is calculated by:
Where \( X_{i}^{t} \) and \( X_{j}^{t} \) are the current position of the fireflies \( X_{i} \) and \( X_{j} \), and \( X_{i}^{t + 1} \) is the ith firefly position of the next generation. The second term is due to attraction. The third term introduces randomization, with \( \alpha \) being the randomization parameter.
The basic steps of the firefly algorithm can be formulated as the pseudo code shown in the Algorithm 1.
The original firefly algorithm is designed for optimization problems with continuous variables. Recently, several binary firefly algorithms were developed to solve discrete problems, such as scheduling, timetabling and combination. Compared with the original firefly algorithm, binary firefly algorithm obeyed similar fundamental principles while redefined distance, attractiveness, or movement of the firefly. In this study, we use a binary firefly algorithm for feature selection with new definitions of distance and movement of a firefly, similar to the approach used in [10].
3 The Proposed Method for Feature Selection and Classification
The feature selection task is a typical combination problem in essence, with the objective of selecting an optimal combination of features from a given feature space. Theoretically, for an n-dimensional feature space, there will be \( 2^{\text{n}} \) possible solutions (NP-hard problem). The use of metaheuristics methods as random selection algorithms, capable of effectively exploring large search spaces, which is usually required in case of feature selection. In this work, a binary firefly algorithm (BFA-SVM) is proposed; where the feature space is explored by a population of fireflies and the SVM classifier is used for evaluating a feature subset. The normalized Hamming distance was used to calculate attractiveness between a pair of fireflies and in order to increase the diversity of fireflies a dynamic mutation operator was introduced. The flowchart of BFA-SVM method is shown in Fig. 1.
3.1 Fireflies Representation and Initialization
To represent the subset of selected features, we chose a binary representation of a solution in the multidimensional search space. Every firefly \( \varvec{x}_{\varvec{i}} \) in the binary firefly algorithm represents a subset of the feature space (i.e. a possible solution for feature selection problem) as an N-dimensional binary array: bit values 1 and 0 represent a selected and unselected feature, respectively. The Initial population of fireflies is generated randomly; the bit positions for each firefly are randomly assigned as 1 or 0.

3.2 Objective Function
The objective function of the BFA-SVM algorithm when searching for the optimal features subset is to maximize the accuracy rate in classifying the testing dataset. This is equivalent to an optimization problem seeking for a maximum solution. The classification rate ACC is calculated using cross-validation with 10-Folds [11]. This measure is calculated by the formula (4):
Where Total correct is the number of examples correctly classified by the SVM classifier, and L is the total number of examples. The classification rate indicates whether the candidate subset permits good class discrimination.
3.3 The Attractiveness of Fireflies
For two fireflies \( X_{i} \) and \( X_{j} \), the distance \( r_{ij} \) is defined based on the similarity ratio of the two fireflies using the normalized Hamming distance of the two position vectors as follow:
Where \( \oplus \) denotes the XOR operation and \( d \) is the positions dimension. The attractiveness β between a pair of fireflies is calculated using the formula (1).
3.4 The Movement of Fireflies
The original firefly algorithm is designed for optimization problems with continuous variables. For the binarization of continuous metaheuristics, there are two main groups of binarization techniques. The first group of techniques allows working with the continuous metaheuristics without operator modifications and includes steps of binarization of the continuous solution after the original continuous iteration. The second group of techniques is called continuous-binary operator transformation; it redefines the algebra of the search space, thereby reformulating the operators [18]. In this work, we use a modification in the movement of a firefly by the reformulation of the formula (2). When a firefly \( X_{i} \) moves to another firefly \( X_{j} \) more attractive, every bit in its representation vector will make a decision to change its value or not. Changing a bit \( X_{i}^{k} \) in firefly \( X_{i} \) is done in two steps: the β-step (attraction) as indicated in the formula (6), which is regulated by the attractiveness β, and the α-step (mutation) were using the formula (7), which is controlled by a parameter α.
β is the probability of a hetero-bit in the moving firefly changes to the corresponding bit in the brighter firefly (0 → 1 or 1 → 0). The parameter α regulates the random moving behavior (mutation) of a bit \( X_{i}^{k} \), and it is calculated in each iteration of the BFA-SVM algorithm by the following formula:
The mutation probability α is high in initial iterations, which makes BFA-SVM focus on exploration. As the number of iteration increases, the mutation probability will decrease, and BFA-SVM will accelerate its converging pace gradually.
4 Experiments
The proposed BFA-SVM algorithm was implemented on a PC with an Intel Core 2 Duo CPU 2.93 GHz, 4 GB of memory and the Windows 7 operating system. The programs are coded in Java language and we have used the LIBSVM package [5] as a library for the SVMs.
4.1 Dataset
To evaluate the performance of the proposed method, we have used 11 medical datasets obtained from the UCI Machine Learning Repository [17]. Table 1 describes the main characteristics of these datasets. The prediction process with the SVMs requires that the dataset must be normalized. The main advantages of such operation are to avoid attributes in greater numeric ranges dominating those in smaller numeric ranges, and to avoid numerical difficulties during the computation step. The range of each feature value is linearly scaled to the range [−1, +1] using the WEKA tools [7].
4.2 Parameter Settings
The parameter values of the proposed algorithm are fixed by an experimental study. After a series of experiments, the different parameters are fixed empirically. The values of each parameter for the proposed method are given in Table 2.
4.3 Numerical Results
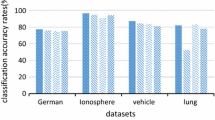
Due to the non-deterministic nature of the proposed method, several executions (20) were considered for each dataset. The minimum value, the maximum value, and the average of the accuracy rate of the classification for each dataset are reported. The best results are in bold font.
Table 3 gives a comparison between the results (mean of accuracy rate) obtained by the use of SVM classifier with default parameters for RBF kernel function, the use of SVM with optimized parameters given with a grid-search method, and the results obtained with by BFA-SVM with optimized parameters. The best results are obtained with BFA-SVM algorithm for all datasets, confirming that the feature selection and optimization of SVM parameters improves significantly the classification accuracy.
-
The SVM with optimized parameters is used as classifier with the algorithm BFA-SVM
-
Method did not use the dataset for test.
-
The best results are in bold.
In order to evaluate the effectiveness of the proposed algorithm BFA-SVM, a comparison of the experimental results obtained by the method with the results of the works cited in [8, 9] is presented in the Table 4, where gives the average (Mean), the best (Max), the worst (Min) values of the classification accuracy, and the standard deviation (Sd) obtained by different methods. In [9], a hybrid search method based on both harmony search algorithm and stochastic local search, combined with a support vector machine (HAS + SVM) is given for feature selection in data classification. And the authors of [8] propose a genetic algorithm (GA) and memetic algorithm (MA) with SVM classifier for feature selection and classification.
As shown in Table 4, BFA-SVM algorithm succeeds in finding the best results for almost the checked datasets compared to HAS + SVM, MA + SVM and GA + SVM methods in term of classification accuracy point of view (in 8 datasets among 11, the BFA-SVM algorithm gives the best classification rate average and for the max value of the classification accuracy is reached in 10 datasets among 11). The small standard deviations of the classification accuracies presented show the consistency of the proposed algorithm. This proves the ability of the proposed algorithm as a good classifier in medical data diagnosis.
5 Conclusion
Health care systems generates vast amount of information and it is accumulated in medical databases, and the manual classification of this data becoming more and more difficult. Therefore, there is an increasing interest in developing automated methods for medical data analysis. In this work we proposed a wrapper method for feature selection and classification of medical dataset based on a Binary firefly algorithm combined with the SVM classifier. The results obtained from tests carried out on several public medical dataset indicate that the proposed BFA-SVM method is competitive with other meta-heuristics (Genetic algorithm, Memetic algorithm and Harmony search algorithm) for the feature selection, and experiments have shown us that the method greatly improves the learning quality and ensures the stability of the generated prediction model. It also reduces the size of the representation space by eliminating noise and redundancy.
As a continuation of this work, it would be desirable to work on the reduction of the computation time by proposing a parallel implementation of the proposed method. It is also possible to use the binary firefly algorithm with other classification algorithms such as neural networks and Naïve Bayes.
References
Almuhaideb, S., El Bachir Menai, M.: Hybrid metaheuristics for medical data classification. In: El-Ghazali, T. (ed.) Hybrid Metaheuristics. Studies in Computational Intelligence, vol. 434, pp. 187–217. Springer, Heidelberg (2013). https://doi.org/10.1007/978-3-642-30671-6_7
Liu, H., Yu, L.: Toward integrating feature selection algorithms for classification and clustering. IEEE Trans. Knowl. Data Eng. 17(4), 491–502 (2005)
Almuhaideb, S., El-Bachir Menai, M.: Impact of preprocessing on medical data classification. Front. Comput. Sci. 10(6), 1082–1102 (2016). https://doi.org/10.1007/s11704-016-5203-5aydin
Vapnik, V.: The Natural of Statistical Learning Theory. Springer, New York (1995). https://doi.org/10.1007/978-1-4757-2440-0
Chang, C.C., Lin, C.J.: LIBSVM: a library for support vector machines. ACM Trans. Intell. Syst. Technol., 2(3), Article 27 (2011). http://www.csie.ntu.edu.tw/~cjlin/libsvm/. Accessed 11 Oct 2017
Yang, X.S.: Nature-Inspired Metaheuristic Algorithms. Luniver Press, UK (2008)
Eibe, F., Mark, A.H., Witten, I.H.: The WEKA Workbench. In: Online Appendix for Data Mining: Practical Machine Learning Tools and Techniques, 4th edn. Morgan Kaufmann (2016)
Nekkaa, M., Boughaci, D.: A memetic algorithm with support vector machine for feature selection and classification. Memetic Comput. 7, 59–73 (2015). https://doi.org/10.1007/s12293-015-0153-2
Nekkaa, M., Boughaci, D.: Hybrid harmony search combined with stochastic local search for feature selection. Neural Process. Lett. 44, 199–220 (2015). https://doi.org/10.1007/s11063-015-9450-5
Zhang, J., Gao, B., Chai, H., Ma, Z., Yang, G.: Identification of DNA-binding proteins using multi-features fusion and binary firefly optimization algorithm. BMC Bioinf. 17, 323 (2016). https://doi.org/10.1186/s12859-016-1201-8
Han, J., Kamber, M.: Data Mining Concepts and Techniques, 2nd edn. Morgan Kaufmann, San Francisco (2006)
Huerta, E.B., Duval, B., Hao, J.-K.: A hybrid GA/SVM approach for gene selection and classification of microarray data. In: Rothlauf, F., Branke, J., Cagnoni, S., Costa, E., Cotta, C., Drechsler, R., Lutton, E., Machado, P., Moore, Jason H., Romero, J., Smith, George D., Squillero, G., Takagi, H. (eds.) EvoWorkshops 2006. LNCS, vol. 3907, pp. 34–44. Springer, Heidelberg (2006). https://doi.org/10.1007/11732242_4
Kecman, V.: Learning and Soft Computing: Support Vector Machines, Neural Networks and Fuzzy Logic Models. The MIT press, London (2001)
Burgers, C.J.C.: A tutorial on support vector machines for pattern recognition. Data Min. Knowl. Discov. 2, 121–167 (1998)
Duval, B., Hao, J.K.: Advances in metaheuristics for gene selection and classification of microarray data. Brief. Bioinf. 11(1), 127–141 (2009)
El Aboudi, N., Benhlima, l.: Review on wrapper feature selection approaches. In: International Conference on Engineering and MIS (ICEMIS), pp. 1–5 (2016)
Lichman, M.: UCI machine learning repository. University of California, School of Information and Computer Science, Irvine (2013). http://archive.ics.uci.edu/ml
Crawford, B., Soto, R., Astorga, G., García, J., Castro, C., Paredes, F.: Putting continuous metaheuristics to work in binary search spaces. Complexity 2017, 19 (2017). https://doi.org/10.1155/2017/8404231
Acknowledgments
The authors would like to thank the developers of the Library for Support Vector Machines (LIBSVM) and the developers of Waikato Environment for Knowledge Analysis (WEKA) for the provision of the open source code.
Author information
Authors and Affiliations
Corresponding author
Editor information
Editors and Affiliations
Rights and permissions
Copyright information
© 2018 IFIP International Federation for Information Processing
About this paper
Cite this paper
Sahmadi, B., Boughaci, D., Rahmani, R., Sissani, N. (2018). A Modified Firefly Algorithm with Support Vector Machine for Medical Data Classification. In: Amine, A., Mouhoub, M., Ait Mohamed, O., Djebbar, B. (eds) Computational Intelligence and Its Applications. CIIA 2018. IFIP Advances in Information and Communication Technology, vol 522. Springer, Cham. https://doi.org/10.1007/978-3-319-89743-1_21
Download citation
DOI: https://doi.org/10.1007/978-3-319-89743-1_21
Published:
Publisher Name: Springer, Cham
Print ISBN: 978-3-319-89742-4
Online ISBN: 978-3-319-89743-1
eBook Packages: Computer ScienceComputer Science (R0)