Abstract
This paper reports the performance of hyperspectral imaging for detecting the adulteration in red-meat species. Line-scanning images are acquired from muscles of lamb, beef, or pork. We consider the states of fresh, frozen, or thawed meat. For each case, packing and unpacking the sample with a transparent bag is considered and evaluated. Meat muscles are defined either as a class of lamb, or as a class of beef or pork. For visualization purposes, fat regions are also considered. We investigate raw spectral features, normalized spectral features, and a combination of spectral and spatial features by using texture properties. Results show that adding texture features to normalized spectral features achieves the best performance, with a 92.8% overall classification accuracy independently of the state of the products. The resulting model provides a high and balanced sensitivity for all classes at all meat stages. The resulting model yields 94% and 90% average sensitivities for detecting lamb or the other meat type, respectively. This paper shows that hyperspectral imaging analysis provides a rapid, reliable, and non-destructive method for detecting the adulteration in red-meat products.
You have full access to this open access chapter, Download conference paper PDF
Similar content being viewed by others
Keywords
- Hyperspectral imaging
- Spectral-spatial features
- Meat classification
- Meat processing
- Adulteration detection
1 Introduction and Background
Adulteration of meat products is an important quality and safety factor of meat (e.g. the addition of another type of meat which may have a lower price compared to the original material).
Traditionally, meat quality and safety attributes are assessed using lab-based methods. Recently, spectroscopic measurements gain increased attention in the field of meat processing, providing optical properties of a single point on the sample surface and mapping those properties onto quality and safety attributes. Such properties can be defined by reflectance or absorbance of light at specific electromagnetic wavelengths [1, 2]. The spectroscopic approach has disadvantages regarding the non-availability of spatial information, the non-inclusion of small-sized objects into the analysis, missing flexibility in measuring particular spectral information, and inability to generate distributions of attributes [3].
Conventional computer vision systems can be used to assess some meat attributes; they can also deal with the spatial information problem not solvable by single-point spectroscopy. Conventional computer vision systems do not provide multi-spectral information; a colour image provides only reflectance values for three particular energy distributions in the visible light (VIS) wavelengths range (identified as Blue, Green, and Red). The studies described in [4,5,6] show applications of colour images for the assessment of food quality.
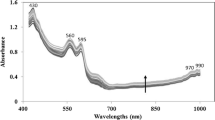
Hyperspectral imaging (HSI) systems aim at a combination of advantages of spectroscopy (i.e. availability of spectral information) with benefits of conventional colour images (i.e. availability of spatial information). Figure 1, left, illustrates an HSI image in spectral [wavelength \(\lambda \)] and spatial [pixel locations (x, y)] coordinates forming a hypercube in this \(xy\lambda \) space. Thus, a spectral imaging system is able to provide quality attributes for spectral information as well as spatial information for the localisation of those spectral data in the sample. Spectral imaging systems facilitate the visualization of objects and the chemical distribution of their components. In general, an HSI system collects information about external attributes (spatial information) and internal attributes (spectral information) as spectral signatures of materials. Figure 1, upper right, shows spectral signatures for four types of materials, namely lamb, beef, and pork muscles, and fat. Spatial and spectral information characterizes physical and chemical features of objects. In general, HSI systems are more reliable than conventional imaging systems or just using spectroscopy technology.
The rest of this paper is structured as follows. Section 2 provides an overview on related techniques for HSI classification and analysis. Section 3 describes the used data set and the used HSI system. Section 4 covers spectral analysis and pattern visualization. Section 5 describes our framework for classifying the red-meat species. After that, results and discussion are given in Sect. 6. Section 7 concludes.
2 Related Work
The classification of HSI data is the main task in many applications, such as in medical applications, remote sensing imagery, or, as considered here, in meat quality processing. In food and meat applications, HSI is considered as being a powerful tool for classifying or predicting attributes related to food quality. In [7], a linear model has been developed to classify the type of lamb muscles. The average of spectra for each sample was used to build a model. The results showed that HSI was able to define the type of muscles of lamb meat samples, while conventional RGB image analysis failed in performing this characterization.
[8] investigates an HSI system for the discrimination between three types of red-meat (lamb, pork, beef) using partial least-squares discriminant analysis (PLS-DA) as a supervised learning model for solving the classification problem. Results showed that PLS-DA performs well in cases of sample-based evaluation, but it provides a misclassification of pixels in cases of pixel-based evaluations. The misclassification of pixels results due to the model being built using the average spectrum of each sample; spatial features are ignored in this case. The system considers the spectral variation in the sample space only, without taking into account the spatial variation in pixel space. In fact, pixels of an HSI image are affected by the source light (light scattering or illumination effects).
In [9], PLS-DA was compared with soft independent modelling of class analogy (SIMCA) for the classification between lamb meat and other types (pork or beef). It was found that PLS-DA performed better than SIMCA, but the method’s performance varied depending on the way samples were presented (i.e. vacuum packed or without packaging).
Understanding the balance between a variation of spectral information in sample space and the effect of light in pixel space, is a real challenge in building any learning model for classifying meat samples. This challenge needs to be addressed in the case of heterogeneous images. For example, detecting any adulteration in pre-packed rolling meat products is of significant importance. In this case, a pixel-wise (i.e. local) prediction is not only more practical but also more reliable than a sample-wise (i.e. global) prediction.
Building the classification model by using the average spectrum of each sample is a common way for collecting the spectral information for each material using an HSI system [7, 8, 11,12,13]. For reducing the effect of light scattering within one image, there are methods commonly used, such as spectral derivatives [8], standard normal variates (SNV) [12], or multiplicative scatter correction (MSC) [12].
In HSI systems, each pixel contributes to the spectral signature of a material. Thus, as a basic strategy, the use of a pixel as a sample of a material might produce a model invariant to local changes within an image. This methodology is commonly applied to hyperspectral imaging for remote sensing applications [10]. It is proposed due to a limitation in resources (i.e. in image data). Thus, taking a pixel as a sample, while considering different samples of material, provides a powerful model with the advantage of considering local changes within an image.
An HSI classification model that uses only spectral features may provide some useful (but not yet fully satisfying) results; in this case, the spatial information is ignored. Spatial features reflect the geometric or topologic structure of an objects interior. In addition, they also provide information about the local variation in spectral data for each pixel. Thus, also taking into account the spatial features requires a conversion of image data from a pixel-oriented into an object-oriented data structure, i.e. defined by image segments, where each segment is defined by some kind of uniformity in spatial information [14].
A method for spectral-spatial feature extraction in HSI applications is given by super-pixel identification. [15] uses simple linear iterative clustering (SLIC) to generate super-pixels of HSI images; the mean of spectral values in each super-pixel is used as an input for an SVM classification model. Then, a linear conditional random-field model is used to compute the final classification map. Following this method, an HSI image is converted into super-pixels based on spectral-spatial information; pixels in each super-pixel have “similar” spectral and spatial features.
Advantages of using super-pixels for extracting spectral features are as follows: (i) averaging over a super-pixel at each wavelength provides a stable spectral signature [15], (ii) a possibility of considering also spatial features [16] (e.g. as known from texture analysis or neighbourhood relationships), and (iii) a potential reduction of computation time in the analysis and prediction phases.
A combination between spectral and spatial features of HIS images are proposed in [16] using a multi-kernel composition of an SVM-RBF. Three types of features are used in this work: the spectrum for each pixel, the average of all pixels inside a generated super-pixel, and a weighted average of eight neighbours for each super-pixel. In [15, 16], experimental results show that the use of super-pixels (or any other image segmentation method) for defining spectral features, potentially improves the performance of the fitted models. However, the use of super-pixels may also reduce the performance if extracted segments are inaccurate. For this reason, the segmentation is a critical step in HSI analysis. Ensemble classification rules, like the methods used in [15, 16], may be more logical and efficient; decisions in these methods are a combination of spectral and spectral-spatial features.
In general, our study aims at exploring the robustness of hyperspectral imaging systems to discriminate between different types of red-meat muscles. The main contributions of this paper are as follows:
-
Investigate the effects of realistic conditions on spectral appearance of fresh red-meat; studied conditions are (1) packing meat into a transparent bag, (2) meat frozen for six hours, and (3) thawing meat after being frozen.
-
Develop a learning model to discriminate one type of meat muscle from the others (e.g. in case of adulteration), for example, identify lamb meat in difference to beef or pork with taking into consideration the conditions mentioned above.
-
Develop a methodology to consider the local variation in both spectral and spatial features by using a method for super-pixel segmentation.
-
Evaluate different types of spectral (e.g. normalization) and spatial (e.g. texture) feature extraction methods.
3 Data Set and HSI System
A collection of three red-meat species were procured from local supermarkets [9]. The total number of procured meat samples is 45, divided as follows: 17 samples of lamb muscle, 13 of beef, and 13 of pork. The samples were randomly partitioned into a calibration set of 30 samples (12 from lamb, 9 from beef, and 9 from pork). The remaining samples were used for evaluation. Pieces of each muscle type were extracted and put into designed frames. Figure 2 shows four of those frames, each having a specific muscle type in one of \(4 \times 4\) cells.
Ground-truth and false-color images for frames 1 to 4 (top to bottom), where FSUP is short for fresh red-meat unpacked, FSP for fresh packed, FRUP for frozen unpacked, FRP for frozen packed, and THUP for frozen-thawed unpacked. In cells of ground truth, F is for fat, L for lamb, B for beef, and P for pork
Line-scanning HSI images were collected from these frames at five different statuses; a set of 20 HSI images was acquired for model calibration when the meat was (1) fresh, (2) freshly packed in a transparent bag, (3) frozen, (4) frozen and packed in a transparent bag, and (5) thawed (after being frozen) and unpacked. These statuses were investigated regarding their effect on the spectrum of red-meat muscles.
The HSI system, which was used in this work, provides a high spectral resolution of 4.9 nm, and it covers a wide range 547.8–1701.2 nm of wavelengths, thus 235 bands across the electromagnetic spectrum. The HSI acquisition system was set up, and the reflectance is calculated following [8, 17]. The reflectance is computed as follows:
This calibrated image reflectance R is obtained from the raw image irradiance \(R_{0}\) by using the dark reference image D and the white reference image W. After reflectance calibration, the first and last five bands of the given 235 bands were removed due to the low SNR in these bands.
4 Spectral Data Analysis and Visualization
For simulating the adulteration in red-meat products, we defined the following problem: Identify lamb muscles in difference to other muscles types (i.e. here beef or pork). Thus, the spectral properties of lamb meat are labelled as being one class (called LAMB), and we have another class spectral properties for both beef and pork (called OTHER). In addition, we also use a class FAT for visualization purposes.
PCA analysis. Left: Mean spectrum of each of the three classes for each of the five statuses; wavelengths between 547.8 nm and 1,701.2 nm versus reflectance values between 0 and 1. Right: Scatter plot for the LAMB and OTHER classes, showing 97.67% of the total variances of the original data; PCA 1 (97.3%) between \(-20\) and +5 versus PCA 4 (0.37%) between \(-2\) and +4
When dealing with HSI images, a challenge is the dimensionality of the image data (their large size). The high dimensionality reflects negatively on data visualization and the analysis of this type of images. Agreeing with other authors [7, 8, 13], we also consider principal component analysis (PCA) as an appropriate model for dealing with the dimensionality of HSI images.
PCA can be used for proving and visualizing the separation between classes of different materials. PCA is used for reducing the dimension of HSI data, thus producing a limited number of images, called score images, sorted by eigenvalue magnitudes from highest to lowest score image (Recall: The highest score images represent the most important spectral information from the original spectral information.) For example, in [16], the first three score images were used as input for a segmentation model for segmenting an HSI image and for extracting spatial features from the resulting segments.
We used the data set as introduced in Sect. 3. The calibration subset was hand-labelled (ground truth) for manually defined image segments; the mean spectrum of each segment was used to represent this segment, for each class of the previously described set of HSI images. Then, PCA was applied on the extracted data set for two reasons: (1) for visualizing the patterns between the pre-defined classes (LAMB and OTHER), and (2) as a pre-processing step for extracting the spatial features from each image for each class.
Figure 3, left, shows the mean spectrum of each class at each status of meat. Clearly, this figure shows that there are significant differences in the mean spectrum for each class. For visually investigating the class separations, Fig. 3, right, shows the distribution of the data in the PCA space where the classes are subdivided into overlapping regions; “overlapping” comes from the data in frozen statuses.
5 Classification Framework
In general, a manual selection of pixels as samples, from each class, is impractical and inefficient for creating a robust learning model because no local changes are considered in this case. For this reason, we propose a super-pixel segmentation to convert the HSI image into a map of super-pixels. The pixels in each super-pixel share “similar” local spectral features. Also, super-pixels reflect local spatial features (e.g. similar texture) in the image.
Then, from each super-pixel we select a limited number of pixels to represent this super-pixel. In this work, the SLIC-superpixel algorithm of [19] is used to generate super-pixels of an HSI image. SLIC was originally proposed for colour images; it is based on measuring the similarity (using the Euclidean distance) in RGB or CIE-LAB space, combined with closeness of spatial coordinates.
Due to the high dimensionality of HSI images, we propose to use SLIC in the PCA space; we use the first (i.e. highest) five score images as input for SLIC; the Euclidean distance is used as similarity measure in the PCA space. Figure 1, lower middle, shows an example of five score images and (lower right) the resulting super-pixel segmentation of an HSI image. The resulting super-pixels are accurate, i.e. without any overlapping between different types of meat. As a classifier, an SVM-RBF algorithm was used for evaluating different types of features; it is also used for obtaining the final classification maps.
Extraction of spectral features. The resulting segmentation map is used for extracting samples (pixels) from each class. Considering all pixels in each segment requires high computing costs. For this reason, and for making the classes balanced, we use the Kennard-Stone (KS) algorithm [20] to select a subset of representative pixels. A challenge in HSI image data is the dependence of the reflectance values from the source light (light scattering). Transforming pixel values (a pixel value is a vector of reflectance values in the considered wavelength range) into a normalized version emphasizes the patterns between the spectral appearance of the classes, and also reduces the effect of the used lighting. By considering the pixel as a vector, there are two ways of normalization. Let \( P{(x,y)} = [u_{1},u_{2},\ldots ,u_{n}]^{T}\) be a pixel in an HSI image at location (x, y).
The first option is to simply convert the pixel-value vector into a unit vector by dividing the values by the \(L_{2}\)-norm \(||P{(x,y)}||_2=\sqrt{{u_{1}}^2+{u_{2}}^2+....+{u_{n}}^2}\) of the vector:
The second option for normalization is a standard normal variation (SNV). In this case, the expected value of pixel values is centred at zero by subtracting the mean \(\mu \) of the vector values, and then scaled into a unit standard deviation by dividing by its standard deviation \(\sigma \):
Practically, an HSI camera is very sensitive to the lighting; typically measured spectral values have many spiking points. These spiking points affect the normalization transformation. The Savitzky-Golay (SG) algorithm [18] is used to reduce the effect of spiking points. The SG method smooths the spectral values by estimating the shape of a group of bands (defined by a window size) by multi-order polynomial fitting. Empirically, we set the window size to 9, with a 2nd-order polynomial fitting to estimate the shape of each pixel before the normalization transformation.
In general, HSI systems produce an enormous amount of spectral information. These cameras are designed to cover a large area of applications such as fruit sorting, medical applications, or, our case, meat processing. Practically, a lot of this information is redundant and not required to accomplish a particular task. Based on this fact, we use recursive feature elimination (RFE) [21, 22] to select a set of “most significant” wavelengths for making a distinction between the lamb muscle and the other muscles. In the RFE procedure, we use a random forest (RF) algorithm to estimate the importance of each wavelength in this classification task. From our PCA analysis, we conclude that the reflectance of each class is strongly affected by the status of the meat. These changes in reflectance value affect the classification results and the importance of various wavelengths. Thus, in this case, we apply an RFE procedure on the data for each status individually (to estimate a set of the most significant wavelengths for each status). Then, a union of all sets is taken with removing any duplication.
Extraction of spectral-spatial features. Texture properties of an image are often used as spatial features in computer vision. A common model for extracting texture features is the gray-level co-occurrence matrix (GLCM); the GLCM supports a statistical methodology for analysing the spatial relationships of adjacent pixels by calculating how often a pair of pixels with the same intensity values occurs in an image; see, e.g., [14]. In [23], Haralick proposes a set of statistical features to represent spatial properties (texture properties) of an image. These features were extracted from the CLCM matrix. We use the following Haralick-features: homogeneity, contrast, inverse difference moment (IDM), entropy, energy, and correlation.
In the case of HSI images, extracting these features is demanding due to having many gray-level images (for wavelengths) inside the hypercube; it is hard to decide which wavelengths represent the texture of objects shown in an image. For dealing with this problem, we used the previous RFE analysis to select six wavelengths that have the highest importance (rank) resulting from the RF model. The selected wavelengths (all in nm) are as follows:
Figure 1, upper right, illustrates that these wavelengths are logical: at these wavelengths we have a significant difference between signatures of the considered different types of red-meat.
For computing the CLCM matrix, we use the SLIC segments by taking a window of \(20 \times 20\) pixels centred in a super-pixel. The center of a super-pixel is defined as the first moment (centre of mass) of all pixels contained in this super-pixel. The selected window is masked by using value 0 to remove any pixel outside the segment border. For avoiding any effect caused by the masked pixels (i.e. the zeros) inside the window, the first row and first column of the CLCM are eliminated, and the selected wavelengths are normalized into the range of 1–255. After that, texture features are computed for each super-pixel at six wavelengths; the total number of the selected features is 36 features for each super-pixel. Then, these sets of features are added to the selected spectral features of pixels inside the considered super-pixel.
6 Experimental Results and Discussion
The data set, described in Sect. 3, is used to build the prediction model. First, the PCA was applied to each image. After that, the first five score images were used as input for the SLIC algorithm. The initial region size of each super-pixel was set to 100 pixels (i.e. \(10 \times 10\)). Then, the resulting segments are labelled by one of the classes (i.e. LAMB, OTHER, or FAT). By using the KS algorithm, from each super-pixel, we select a limited number of pixels (11 for lamb and 9 for the other cases) to represent the super-pixel. Then, a data set was built from these pixels.
We investigated the following feature vectors: Raw spectral, normalized spectrum by L\(_2\) norm (L \(_2\) -norm), normalized spectrum by L\(_2\) norm with texture (L \(_2\) -norm-texture), SNV normalization (SNV-norm), and SNV normalization with texture (SNV-norm-texture). In the case of raw spectral, raw reflectance features (by applying Eq. (1)) were considered, where the total number of features is 225, while in the normalization case, the raw spectral data were smoothed, then processed by applying Eq. (1), and then normalised using either \({P}^\circ \) as in Eq. (2) for the L\(_2\)-norm, or \(\overline{P}\) as in Eq. (3) for the SNV-norm.
For reducing the dimensionality of the features, only optimal features (wavelengths) of the RF model were chosen (93 and 103 features for the L\(_2\)-norm and the SNV-norm, respectively). As described above, we select a set of 36 spatial features to represent the texture of each super-pixel. Thus, all the pixels which belong to the same super-pixel have the same spatial features. Then, these spatial features were added to the feature vectors in the case of L\(_2\)-norm and SNV-norm.
Visualization results (classification maps) of the proposed feature vectors. The colors Red, Green, and Blue represent classes LAMB, OTHER (beef or pork), and FAT, respectively. Top to bottom: FSUP, FSP, FRUP, FRP; all four rows for HSI of fresh and frozen (packed and unpacked) red-meat, and, in the row at the bottom, THUP for HSI of fresh thawed unpacked red-meat (Color figure online)
We use the SVM-RBF algorithm for performing classification. For model assessment, we use a 10-fold cross-validation with grid search [24] for hyper-parameterFootnote 1 selection. For evaluating the resulting models, a new set of samples (6 for lamb, 7 for beef, and 6 for pork) were evaluated and analyzed. In more detail, these samples were prepared and put in a private frame for imaging and simulating the situation of mixing different types of red-meat. Then, HSI images were captured for the meat at the following stages: two images for fresh meat (packed and unpacked), another two after having the meat frozen for six hours (packed and unpacked), and one image after thawing the frozen samples. The first column of Fig. 4 shows false color images of the HSI images which were used to evaluate the proposed features. The second column shows the selected regions for quantitative assessment (ground-truth). Some fat regions were eliminated from the assessment due to the fact that these regions are skinny fat, and this kind of fat was not considered during the model calibration.
The evaluation results show that HSI systems are robust tools for detecting the adulteration in red-meat products. Moreover, this robustness is invariant to the state of the products (e.g. fresh, packed, frozen, or frozen-thawed).
In case of sample-wise evaluation, visually, the proposed feature vector (SNV-norm-texture) provides a very accurate performance, where all samples of all types of meat are classified successfully. But a main objective of this paper was to evaluate the proposed features in a way of pixel-wise evaluation.
Table 1 shows pixel-wise quantitative performance results for each feature vector. In all cases, the proposed spectral normalization methods enhance the accuracy compared to a plain-spectral method.
The results show that raw spectral data are strongly affected by the state of the meat. The best overall accuracy was achieved for the state of fresh meat. However, the accuracy significantly decreased for the other statuses; this observation clearly appeared from our PCA analysis. The changes in accuracy document the need that other features need to be used. Our goal is to provide a model with a high stability between the sensitivity and precision of all classes.
Compared to raw spectral data, the enhancement given by the proposed method is even more obvious for the other meat states. For example, in case of FSP, the accuracy increased from 83.3% to 90.3%. In general, raw spectral features produced a non-stable model; for example, the sensitivity of classes (LAMB, and OTHER) significantly change from state to state. Also, there is a gap in the sensitivity of the classes at all the statuses except the FSUP status while normalizing the spectrum provides a more stable performance. The gap between the sensitivity of the classes is significantly reduced.
Table 2 shows the results (sensitivity, precision, and overall accuracy) on average for all meat statuses. On average, the SNV normalization outperforms the L\(_2\) normalization where the mean overall accuracy of all meat statuses are 87.2% and 91.4% for L\(_2\)-norm and SNV-norm, respectively.
As expected, by adding spatial features (e.g. texture) we improve the accuracy and the stability of the proposed spectral normalization methods. In the case of the L\(_2\)-norm, the spatial features increase mean accuracy from 87.2% to 89.8%; in the case of SNV-norm, they increase mean accuracy from 91.3% to 92.8%.
By adding the spatial features we significantly improved the accuracy for meat statuses FSP and THUP. For example, in THUP, the accuracy jumped from 86.86% (raw spectral) to 95.5% (SNV-norm-texture), while there is no significant enhancement in the accuracy of statuses FSUP, FRUP, and FRP.
Also, adding texture reflects on the stability of the model; the gap in sensitivity and precision between the class LAMB and class OTHER decreases. This suggests that the model resulting from SNV with texture is the best and most efficient model in the set of considered models. Figure 4 shows the resulting classification map of each feature vector for each meat status. The last column shows results for the best-achieved accuracy which occurred when texture properties were added to the SNV-normalized spectral features.
7 Conclusions
Adulteration of red-meat products is a growing concern to the industry. This study investigates the use of HSI to detect adulteration independently of the state of the products (fresh, packed, frozen, or frozen and thawed). To achieve this goal, we investigated different types of spectral and spatial features. The quantitative performance analysis shows that SNV normalization with texture features produces a stable model, fairly invariant to the red-meat status with 92.8% average overall accuracy. The results show that packing the sample into a transparent bag did not affect the spectral response of that sample if it is packed tightly. Lamb meat is detected successfully without any misclassification of pieces with high sensitivity in the case of pixel-wise evaluation while the classification results of beef or pork are affected by the status of the meat, especially in the frozen status; here is space for improvements.
Notes
- 1.
Hyper-parameters are parameters that are not directly learnt within estimators.
References
Bock, J., Connelly, R.: Innovative uses of near-infrared spectroscopy in food processing. J. Food Sci. 73, 91–98 (2008)
Cen, H., He, Y.: Theory and application of near-infrared reflectance spectroscopy in determination of food quality. Trends Food Sci. Technol. 18, 72–83 (2007)
Elmasry, G., Kamruzzaman, M., Sun, D.-W., Allen, P.: Principles and applications of hyperspectral imaging in quality evaluation of agro-food products: a review. Crit. Rev. Food Sci. Nutr. 52, 999–1023 (2012)
Du, C., Sun, D.: Comparison of three methods for classification of pizza topping using different colour space transformations. J. Food Eng. 68, 277–287 (2005)
Zheng, C., Sun, D., Zheng, L.: Recent developments and applications of image features for food quality evaluation and inspection: a review. Trends Food Sci. Technol. 17, 642–655 (2006)
Wu, D., Sun, D.: Colour measurements by computer vision for food quality control: a review. Trends Food Sci. Technol. 29, 5–20 (2013)
Kamruzzaman, M., Elmasry, G., Sun, D., Allen, P.: Application of NIR hyperspectral imaging for discrimination of lamb muscles. J. Food Eng. 104, 332–340 (2012)
Kamruzzaman, M., Barbin, D., Elmasry, G., Sun, D., Allen, P.: Potential of hyperspectral imaging and pattern recognition for categorization and authentication of red meat. Innov. Food Sci. Emerg. Technol. 104, 332–340 (2012)
Karrer, A., Stuart, A, Craigie, C., Taukiri, K., Reis, M.M.: Detection of adulteration in meat product using of hyperspectral imaging. In: Proceedings of Chemometrics Analytical Chemistry, p. 177 (2016)
Ghamisi, P., Couceiro, M., Benediktsson, J.: Integration of segmentation techniques for classification of hyperspectral images. IEEE Geosci. Remote Sens. Lett. 11, 342–346 (2014)
Ropodi, A., Pavlidis, D., Moharebb, F., Panagou, E., Nychas, G.: Multispectral image analysis approach to detect adulteration of beef and pork in raw meats. Food Res. Int. 67, 12–18 (2015)
Kamruzzaman, M., Makino, Y., Oshita, S.: Rapid and Non-destructive detection of chicken adulteration in minced beef using visible near-infrared hyperspectral imaging and machine learning. J. Food Eng. 170, 8–15 (2016)
Sanz, J., Fernandes, A., Barrenechea, E., Silva, S., Santos, V., Goncalves, N., Paternain, D., Jurio, A., Melo-Pinto, P.: Lamb muscle discrimination using hyperspectral imaging comparison of various machine learning algorithms. J. Food Eng. 174, 92–100 (2016)
Klette, K.: Concise Computer Vision. UTCS. Springer, London (2014). https://doi.org/10.1007/978-1-4471-6320-6
Hu, Y., Monteiro, S., Saber, E.: Super pixel based classification using conditional random fields for hyperspectral images. In: Proceedings of IEEE International Conference on Image Processing, pp. 2202–2205 (2016)
Fang, L., Li, S., Duan, W.: Classification of hyperspectral images by exploiting spectral-spatial information of superpixel via multiple kernels. IEEE Trans. Geosci. Remote Sens. 53, 6663–6674 (2015)
Burger, J., Geladi, P.: Hyperspectral NIR image regression part I: calibration and correction. J. Chemom. 19, 355–363 (2005)
Savitzky, A., Golay, M.: Smoothing and differentiation of data by simplified least squares procedures. Anal. Chem. 36, 1627–1639 (1964)
Achanta, R., Shaji, A., Smith, K., Lucchi, A., Fua, P., Susstrunk, S.: SLIC superpixels compared to state-of-the-art superpixel methods. IEEE Trans. Pattern Anal. Mach. Intell. 34(11), 2274–2282 (2012)
Kennard, R., Stone, L.: Computer aided design of experiments. Technometrics 11(1), 137–148 (1969)
Ambroise, C., McLachlan, G.: Selection bias in gene extraction on the basis of microarray gene-expression data. Proc. Natl. Acad. Sci. 99(10), 6562–6566 (2002)
Svetnik, V., Liaw, A., Tong, C., Wang, T.: Application of Breiman’s random forest to modeling structure-activity relationships of pharmaceutical molecules. In: Roli, F., Kittler, J., Windeatt, T. (eds.) MCS 2004. LNCS, vol. 3077, pp. 334–343. Springer, Heidelberg (2004). https://doi.org/10.1007/978-3-540-25966-4_33
Haralick, R.: Statistical and structural approaches to texture. Proc. IEEE 67(5), 786–804 (1979)
Hsu, C.W., Chang, C.C., Lin, C.J.: A practical guide to support vector classification. Technical report, National Taiwan University (2003, last update 2016)
Acknowledgments
Authors appreciate funding by the AgResearch Core Fund and the Ministry of Business, Innovation and Employment, New Zealand. Authors also acknowledge conference support by Auckland University of Technology, the School of Engineering, Computer and Mathematical Sciences.
Author information
Authors and Affiliations
Corresponding authors
Editor information
Editors and Affiliations
Rights and permissions
Copyright information
© 2018 Springer International Publishing AG, part of Springer Nature
About this paper
Cite this paper
Al-Sarayreh, M., Reis, M.M., Yan, W.Q., Klette, R. (2018). Detection of Adulteration in Red Meat Species Using Hyperspectral Imaging. In: Paul, M., Hitoshi, C., Huang, Q. (eds) Image and Video Technology. PSIVT 2017. Lecture Notes in Computer Science(), vol 10749. Springer, Cham. https://doi.org/10.1007/978-3-319-75786-5_16
Download citation
DOI: https://doi.org/10.1007/978-3-319-75786-5_16
Published:
Publisher Name: Springer, Cham
Print ISBN: 978-3-319-75785-8
Online ISBN: 978-3-319-75786-5
eBook Packages: Computer ScienceComputer Science (R0)