Abstract
Proprotein convertases (PCs), also known as eukaryotic subtilases, are a group of serine proteases comprising furin (PACE), PC1 (PC3), PC2, PC4, PACE4, PC5 (PC6), and PC7 (LPC, PC8) that generate bioactive proteins and peptides, such as hormones, receptors, and growth factors by cleaving precursor proteins at multibasic motifs. Two other family members, SKI-1/S1P and PCSK9, cleave regulator proteins involved in cholesterol and fatty acid homeostasis at nonbasic peptide bonds. Furin is ubiquitous in eukaryotic tissues and cells. PACE4, PC5, and PC7 are also widespread, whereas the expression of the other PCs is more restricted. PCs are synthesized as multi-segmented zymogens which are autocatalytically activated. The prodomains have regulatory and inhibitory functions. The catalytic domains are the most conserved domains among the PCs. The architecture of the catalytic active furin domain is known in different binding states. The C-terminal parts of the PCs differ in length and structure and contain encoded peptide signatures guiding the PCs to the subcellular destinations on the secretory pathways: SKI-1/S1P to the cis-Golgi, furin, PC5B, and PC7 to the TGN region but also to the plasma membrane. PACE4, PC5A, and PCSK9 are attached at the cell surface. Truncated, soluble furin and SKI-1/S1P, as well as PC1 and PC2, are released into the extracellular matrix. Many enveloped viruses are activated by furin and furin-like PCs and arenaviruses and a few bunyaviruses by SKI-1/S1P. The PCs cleave the viral fusion glycoprotein to trigger fusion of viral envelopes with cellular membranes to deliver the viral genome into host cells. Cleavage by PCs, occasionally in concert with other endoproteases, enables conformational changes in the viral membrane proteins needed for correct oligomerization of glycoprotein spikes and their effective incorporation into virions. Mutational alterations of PC cleavage sites can reduce the fusion potential of viral surface proteins and thus facilitate the development of secure live attenuated vaccines. Alternatively, agents preventing cleavage of viral surface (glyco)proteins block fusion capacity and multicyclic virus replications. PC inhibitors are suggested as promising antiviral drugs for quite a number of viruses causing severe infections.
You have full access to this open access chapter, Download chapter PDF
Similar content being viewed by others
Keywords
- Furin
- PC1/3
- PC2
- PACE4
- PC5/6
- PC7/8
- SKI-1/S1P
- Subcellular localization and trafficking
- PCs structure and biosynthesis
- Fusion proteins
- Biologically active peptides
- Glycoprotein trimerization and incorporation
- Protease activation mutants
1 Introduction
Many biologically active proteins are synthesized as larger inactive precursors (proproteins). Posttranslational processing by limited proteolysis of the precursors is a mechanism generating active proteins and peptides that enables cells to regulate many vital processes. In general, processing occurs when the proteins are transferred in the secretory pathway from the endoplasmic reticulum (ER) to other cell compartments and to the cell surface. The precursors are frequently cleaved at amino acid motifs containing single or multiple arginine or lysine residues. For a long time, little was known about the activation proteases. The situation changed when the genomes of a wide variety of organisms were decoded. Roughly 600 different proteases have been identified in each genome allowing expression and functional studies. Differing in mammalian species, roughly 200 are serine proteases that can be divided into about 20 subgroups based on structure, enzymatic properties, and physiological functions. The available evidence indicates that several of these are involved in processing at basic cleavage sites (Puente et al. 2003).
Proinsulin was the first identified precursor polypeptide that contains two dibasic cleavage motifs, arginine-arginine and lysine-arginine (Steiner et al. 1967). Comparison of mature insulin with its precursor suggested that a dibasic-specific endoprotease and a B-type-specific carboxypeptidase are responsible for correct processing, and this concept was further substantiated by the observation that insulin was obtained when the activating enzymes were surrogated by trypsin and carboxypeptidase B in vitro (Kemmler et al. 1971). Subsequently, the number of inactive proproteins with presumed and meanwhile ascertained di- and multibasic cleavage motifs continuously increased. The list started with pro-opiomelanocortin (POMC) , proparathyroid hormone , proalbumin , pro-beta-secretase , pro-nerve growth factor , and the proproteins and propeptides of ß-lipotropic hormone, ß-melanocyte-stimulating hormone (β-MSH), γ-lipotropin (γ-LPH), β-lipotropin (β-LPH), and von Willebrand factor and is steadily upgraded (Barr 1991; Seidah 2011). A large number of precursor proteins are cleaved at K K↓, R K↓, K R↓, or R R↓, which are frequently combined in the consensus sequences (K/R)×n(K/R)↓, where a variable number (n = 0,2,4,6) of basic and nonbasic amino acids separates the flanking basic amino acids.
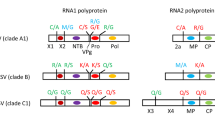
The first authentic proprotein converting enzyme identified was the calcium-dependent subtilisin-like protease kexin encoded by the kex2 gene of Saccharomyces cerevisiae that cleaves yeast and mammalian proteins and peptides at dibasic peptide sites (Achstetter and Wolf 1985; Fuller et al. 1989a; Julius et al. 1984; Thomas et al. 1988). The first mammalian orthologue that possesses the capacity for such a proprotein cleavage was furin (Fuller et al. 1989b; Bresnahan et al. 1990; Wise et al. 1990; Hatsuzawa et al. 1990; Misumi et al. 1990b; van de Ven et al. 1990). Furin cleaves many precursor proteins at the C-terminal end of the motif RX(K/R)R↓ (Barr 1991; Nakayama 1997). Later additional closely related subtilisin-/kexin-like serine proteases were identified and designated proprotein convertases (PCs) (Seidah 2011). The PC family now contains nine members: furin, PC1/3, PC2, PC4, PACE4, PC5/6 (further on PC5), PC7, SKI-1/S1P, and PCSK9 (Table 9.1). The first seven PCs cleave substrates C-terminally at arginine of multibasic recognition motifs. The last two PCs, SKI-1/S1P and PCSK9, recognize nonbasic scissile peptide bonds. The subtilisin-/kexin-like isoenzyme (SKI-1), also known as site-1 protease (S1P), cleaves proproteins at the motif RX(L/I/V)X↓, where X presents any amino acid. The neural apoptosis-regulated convertase 1 (NARC-1), also designated as proprotein convertase subtilisin/kexin type 9 (PCSK9), is autocatalytically cleaved at the amino acid motif VFAQ↓SIP and does not cleave other proproteins in trans but has substrate binding and signaling functions (Seidah et al. 2014). The main cleavage site specificities of the individual PCs are shown in Table 9.1. There are several excellent reviews in which the PC field has been described in detail (Artenstein and Opal 2011; Nakayama 1997; Seidah 2011; Seidah et al. 2013; Seidah and Prat 2002, 2012; Steiner 1998; Thomas 2002).
Many cell proproteins , viral envelope glycoproteins , and bacterial toxins exhibit multibasic cleavage sites (reviewed by Klenk and Garten 1994; Gordon and Leppla 1994) (Tables 9.2 and 9.3). Multibasic cleavage was shown first with the hemagglutinin of fowl plague virus (FPV), a highly pathogenic avian influenza virus (HPAIV), and the fusion protein of virulent Newcastle disease virus (NDV) strains (Bosch et al. 1981; Garten et al. 1981, 1982; Toyoda et al. 1987; Nagai 1995). Cleavage of these glycoproteins in practically all cells allows rapid virus spread in the infected host and proved to be a major determinant for the high pathogenicity of these viruses. The first hint for the nature of the activating host proteases came from the observation that activation of FPV was calcium-dependent, a characteristic feature of PCs (Klenk et al. 1984). The final proof was provided by the identification of furin as the enzyme activating the hemagglutinin of HPAIV and the HIV env glycoprotein (Stieneke-Gröber et al. 1992; Hallenberger et al. 1992).
This chapter gives an overview on furin and the other members of the PC family with a focus on those involved in the life cycle of viruses. The structure and function of the proteases and their biosynthesis, subcellular trafficking, and localization in cells will be shown, as well as their occurrence in specific cell types, tissues, and organisms. PCs have a multifunctional role in virus infection . The main function is the proteolytic activation of fusion-competent surface proteins of enveloped viruses required for the delivery of the viral genome into host cells. Beyond that, PCs are responsible for conformation changes of proteins in virus assembly, for receptor recognition , and for release of pathogenicity factors . PCs together with other proteases are involved in complex cleavage patterns of viral surface proteins. The physiological roles of PCs, especially in embryonic development , as revealed by knockdown systems and specific inhibitory agents will also be discussed. Finally, light will be drawn on the use of protease activation mutants for vaccine design and on the use of protease inhibitors for antiviral therapy .
2 Structure and Biosynthesis of PCs
All PCs are synthesized as multi-segmented pro-precursors which start with an N-terminal signal peptide; continue with a prodomain (prosegment, propeptide), a catalytic domain, and a P-domain (middle domain); and complete the PC ectodomain with variable C-terminal domains (Fig. 9.1). The convertases PC1/3, PC2, PC4, PC5A, PACE4, and PCSK9 are expressed as soluble- or membrane-attached PCs. Furin, PC5B, PC7, and SKI-1/S1P possess a transmembrane anchor domain and a C-terminal cytoplasmic domain. Furin and SKI-1/S1P can be cleaved at a distinct peptide bond in the ectodomain, and thus both convertases exist also as soluble furin (sfurin) and soluble SKI-1/S1P (sSKI-1).
Domain structure of proprotein convertases. Individual domains are illustrated by colored bars, and the total lengths of the human PCs are indicated by the number of amino acids. D, H, N, and S are catalytic active amino acids of the catalytic triad; N contributes to the oxyanion hole. Autocatalytic cleavage sites between and within pro- and catalytic domains of all PCs and the cleavage sites generating soluble forms of furin and SKI-1/S1P are indicated by arrows. C-terminal gray boxes indicate peptide sequences required for cell membrane attachment. The percentage numbers indicate amino acid identity of the catalytic domains relative to furin (Seidah and Prat 2012; Nakayama 1997)
The prodomain (~ 80 amino acids) of the precursor PCs acts as an intramolecular chaperone that guides the folding and activation of the pro-convertases. Simultaneously, the prodomain and/or their autocatalytically split-off fragments function as PC inhibitors. The catalytic domain is the most conserved domain and covers about 340 amino acids with the reactive amino acids aspartic acid (D), histidine (H), and serine (S) in subtilisin-like arrangement and a specific asparagine (N) residue forming an oxyanion hole (Fig. 9.2). The other domains of the convertases are quite divergent in size, sequence homology, and function. The adjacent P-domain confers structural stability and regulates the enzymatic activity. Furin, PC5, and PACE4 contain a conserved cysteine-rich domain (CRD) (Nakayama 1997). CRD functions as a cell surface anchor and interacts with the tissue inhibitors of metalloproteinases (TIMPs) (Nour et al. 2005). The transmembrane domain of furin, PC5B, PC7, and SKI-1/S1P anchors the cytoplasmic domains in the membrane system of the constitutive exocytic pathway. The cytoplasmic domains possess several intrinsic signals determining the residence for each PCs in specific compartments of the constitutive secretory pathway.
Crystal structure of the inhibitor Dec-Arg-Val-Lys-Arg-CMK in complex with mouse furin. The inhibitor is shown in sticks with carbon atoms in yellow, nitrogen in blue, and oxygen in red; the P1–P4 residues and the N-terminal decanoyl group are labeled. The catalytic domain of furin is presented with its surface in green and the P-domain in light blue (Henrich et al. 2003)
2.1 Structure of the Catalytic Domain
Furin is the prototype and by now the best-characterized member of the PC family. Because of difficulties in purifying the enzyme in sufficient amounts and quality, initial studies on the catalytic domain of human furin were based on homology modeling using the crystal structures of the related serine proteinase subtilisin BPN and thermitase of bacterial origin (Siezen et al. 1994). Than and colleagues succeeded in establishing the first crystal structure of a truncated form of furin which was expressed in sufficient quantities and purified from mammalian cells as enzymatically active molecule by Lindberg and coworkers (Henrich et al. 2003; Fig. 9.2). The catalytic pocket carried the covalently bound inhibitor decanoyl-Arg-Val-Lys-Arg-chloromethylketone (Dec-RVKR-CMK), which belongs to the first small peptidyl inhibitors designed for furin (Garten et al. 1989, 1994). This approach revealed an arrangement of highly negatively charged amino acids, i.e., aspartic and glutamic acids, around the catalytic pocket which explains the binding of substrates with the preferential basic amino acids in distinct positions.
As found with all other members of the PC family, the P-domain of furin stabilizes the catalytic domain (Zhou et al. 1998; Than et al. 2005). Detailed knowledge on enzyme-substrate binding properties was achieved by crystallographic analysis of furin loaded with inhibitors or bound to an inhibitory antibody (Dahms et al. 2014, 2016a, b). Very recently, the structure of an unliganded form of furin was determined (Dahms et al. 2016b), where furin exists in a so-called off state, which is incompatible with substrate binding. Moreover, two structures of ethylenediaminetetraacetic acid (EDTA)-treated forms of furin have been determined, one unliganded and the second in complex with a substrate analogue inhibitor. These studies revealed different affinities of three calcium ions, because only two distinct calcium ions have been removed by EDTA treatment. The transition from the off to the on state is triggered by ligand binding and appears to be the precondition for the preferential recognition of the four-residue sequence motif of furin substrates. Moreover, the comparison with EDTA-treated furin structures revealed that the ligation by the presence of calcium influences the active-site geometry and thus modulates furin activity (Dahms et al. 2016b). Recently, furin was complexed with a small non-substrate-like non-peptidic inhibitor which induces structural distortions of the active site of the enzyme (Dahms et al. 2017). Based on these observations taken together, the high substrate specificity of furin can be explained by conformational changes triggered by binding of substrate and calcium. The detailed structural studies also allowed the rational design of novel inhibitors of furin (see Chap. 11).
The only x-ray structure of another PC is that of human PCSK9 which provides detailed insight into its exceptional biochemical characteristics and biological function (Cunningham et al. 2007; Piper et al. 2007). The full-length PCSK9 precursor (amino acids 31–692) was used for crystallization, and the architecture of a very high portion of the polypeptide chain (amino acids 61–683) was determined at 2.3 Å resolution (Fig. 9.3). The structure model comprises nearly the complete prodomain with only two smaller disorders in both terminal peptide regions remaining unsolved. There is a strong interaction between the prodomain and the catalytic domain which explains the inaccessibility of any extern substrate to the catalytic cleft of PCSK9. The C-terminal domain (termed V domain) of this enzyme has a unique structure that allows binding to the low-density lipoprotein receptor (LDLR), which is a target of PCSK9.
Structure of PCSK9 in its autocatalytically cleaved form. The prodomain is shown in cartoon style (cyan) with its C-terminal heptapeptide segment 146DSSVFAQ152 as sticks with carbon atoms in orange, which is bound and inhibits the active site. The catalytic domain is shown with a transparent surface in yellow; the residues of the catalytic triad (S386, H226, and D186) are labeled and provided as sticks (PDB: 2P4E; Cunningham et al. 2007)
Three-dimensional structures of other human PCs are currently not available, but their catalytic domains have been modeled based on the crystal structure of furin . The catalytic domains of PC4, PACE4, PC5/6, and furin resemble each other, whereas those of PC1/3, PC2, and PC7 are less similar (Henrich et al. 2005).
2.2 Structure and Function of the Prodomain
The three-dimensional prodomain structures were solved with PCSK9 and PC1/3. The prodomain of PCSK9 was determined by x-ray crystallography, that of PC1/3 by NMR spectroscopy. The prodomain shows a well-ordered core consisting of a four-stranded antiparallel β-sheet with two α-helices packed against one side of this sheet (Tangrea et al. 2002; Cunningham et al. 2007). Both prodomain structures are similar assuming that all other PC prodomains are structurally closely related. The prodomain is an independent separate domain which possesses three functions: (1) it masks the catalytic domain, (2) it blocks the enzyme activity a priori, and (3) it regulates the dissociation of autocatalytic cleavage fragments at distinct stages on the secretory pathway in a pH-dependent manner. The mechanism how the PCs are differentially processed in space and time has been elucidated in recent years. The concept of pH sensors is based on the histidine content present in the prodomains of the PCs traveling along the pH gradient of the secretory pathway. Histidine-69 (human furin) is conserved in the prodomains of all PCs and functions as the master histidine-encoded pH sensor which regulates enzyme activation at distinct pH values depending on the numbers of the residual histidine residues present in the prodomain. The prodomain of furin is activated at pH ~6.5 within the trans-Golgi network (TGN), whereas the PC1/3 prodomain is activated at pH ~5.5 within the dense-core secretory granules, whereby the prodomain of furin has twice the content of histidine residues in comparison with the prodomain of PC1/3. The correlation of pH-dependent activation and histidine content was demonstrated by swapping the prodomains between both closely related PC family members (Dillon et al. 2012; Shinde and Thomas 2011; Williamson et al. 2013).
2.3 Biosynthesis, Maturation, and Subcellular Localization of PCs
Furin, PC7, PC5B, and SKI-1 are synthesized as class I membrane proteins and the other PCs as soluble secretory proteins (Figs. 9.1 and 9.4). The preproproteins start with the signal peptides which translocate the nascent polypeptide chains from the cytoplasm into the lumen of the ER where the signal peptides are cotranslationally removed from the adjacent prosegment (prodomain). As mentioned above, the newly synthesized prodomains facilitate the folding of the polypeptide chains and block the proteolytic activity of the proPCs until the prodomain fragments are split off and removed at distinct stages of the secretory pathway in a pH-dependent manner. PC1/3 and PC2 become active in secretory granules, furin, PACE4, and PC5B in the TGN region, and PC7 is accumulated in an active form in a separate vesicular structure close to the TGN. SKI-1/S1P is functionally active in the endoplasmic reticulum/Golgi (ER/Golgi) and in the post-Golgi/TGN area (Brown and Goldstein 1999; Lenz et al. 2001; Beyer et al. 2003). PC2 requires the neuroendocrine bifunctional chaperone 7B2 for egress of the ER; its N-terminal domain facilitates the maturation of proPC2, and its C-terminal peptide simultaneously functions as a potent inhibitor (Braks and Martens 1994; Zhu and Lindberg 1995). PC4, PC7, and PCSK9 are processed at a single cleavage site at the boundary between the prodomain and catalytic domain. The other convertases have a second cleavage site, except for SKI-1/S1P which possesses a third cleavage site (Burri et al. 2012a; da Palma et al. 2014; Fig. 9.1). SKI-1/S1P becomes enzymatically active in trans before all prodomain fragments are dissociated from the enzyme (Elagoz et al. 2002; da Palma et al. 2016).
Subcellular trafficking and residences of PCs in a virus-infected cell. The scheme shows the initial infection steps of an enveloped virus with glycoprotein spikes (blue) and the biosynthetic pathway of virus spikes from the endoplasmic reticulum (ER) to the plasma membrane where assembly and release of the virus take place. PCs relevant for virus maturation (magenta) are shown on the exocytic pathway from ER via the Golgi apparatus, the trans-Golgi network (TGN), and the secretory vesicles (SV) to the plasma membrane. PCs recycle via endosomes to their residences: furin and PC5B and PACE4 accumulate in the TGN. PC7 is enriched and enzymatically active in sialyltransferase-deficient vesicles (SDV) and in minor quantities directly exported to the plasma membrane circumventing the Golgi/TGN complex. SKI-1/S1P is mainly active in the cis-Golgi region. The furin ectodomain is cut off by an unknown protease, and SKI1/S1P is probably released by autocatalysis. Soluble enzymatically active furin (sfurin) and soluble sSKI-1 are released into the extracellular matrix; PACE4, PCSK9, and PC5A interact with tissue inhibitors of metalloproteases (TIMPs) and form tertiary complexes with heparan sulfate proteoglycans (HSPGs) ready for cleavage of extracellular substrates. PC1/3, PC2, PC5A, and PCSK9 (black) are most probably irrelevant for processing of viral proteins
The furin-like PCs furin, PACE4, PC5B, and PC7 are trafficking along the constitutive secretory pathway from the ER to the plasma membrane. They show the highest activity in the TGN region sharing the common route with their substrates (Fig. 9.4). PCs recycle to the TGN and to the cis-Golgi. A minor portion of PC7 is routed form the ER directly to the plasma membrane. PC5 exists in two forms, soluble PC5A and membrane-anchored PC5B, due to different gene splicing. Both forms diverge from the TGN in different routes; PC5A migrates via the regulated pathway passing the secretory granules to the plasma membrane where the enzymatically active convertase is tethered to heparan sulfate proteoglycans like PACE4 (Nour et al. 2005). PC7 is arrested as an active integral protease in the TGN-derived sialyltransferase-deficient vesicles (SDV) (Wouters et al. 1998; Declercq et al. 2017). SKI-1/S1P resides preferentially in the late ER and cis-Golgi region where it cleaves the sterol regulatory element-binding protein (SREBP) among other substrates (Brown and Goldstein 1999). PCSK9 is transported via the secretory pathway and secreted in the medium outside from the cell. PC1/3 and PC2 are transported along the regulatory pathway to the dense-core secretory granules, from where they are released by neuronal or hormonal stimuli into the cellular environment (Thomas 2002; Seidah and Prat 2002; Lee and Lindberg 2008).
Subcellular localization of the individual PCs is determined by intrinsic sorting signals (Fig. 9.5). Furin was one of the first molecules shown to accumulate in the TGN under steady-state conditions governed by a molecular address in the cytoplasmic domain (Schäfer et al. 1995; Bosshart et al. 1994; Takahashi et al. 1995). The address is necessary and sufficient for TGN localization and for recycling by the clathrin endocytosis pathway through endosomes. It consists of several destination-determining signals in the form of short peptide sections, which together are necessary for an efficient accumulation of furin in the TGN. They include (1) the acidic signal CPSDSEEDEG783 containing two casein kinase II (CKII) phosphorylation sites, (2) the internalization signal YKGL765, (3) a leucine-isoleucine signal LI760, and (4) the signal F790 (Vey et al. 1994; Schäfer et al. 1995; Molloy et al. 1999; Teuchert et al. 1999a, b; Stroh et al. 1999; Voorhees et al. 1995; Thomas 2002).
Sorting signals of membrane-anchored PCs. Cytoplasmic domains of furin, PC5B, and PC7 are shown. Furin accumulates in the TGN. Furin endocytosis signals are a di-leucine signal (LI), a YXXL signal (YKGL), and a single phenylalanine signal (F). In concert with an acidic peptide containing two phosphorylation sites at serine, they are responsible for TGN localization. Similar peptide elements are found in the cytoplasmic domain of PC5B indicating the TGN destination. PC7 accumulates in TGN-derived sialyltransferase-deficient vesicles (SDV) using a localization signal composed of the peptide segment PLC, the basic peptide HRSRKAK, and two palmitoylated cysteine residues
The cytoplasmic domain of PC5B contains signal elements homologous to furin: the motif YXXL/I, acidic peptide stretches containing serine residues as potential casein kinase II phosphorylation sites, and dileucine motives (Fig. 9.5). Therefore, the transport pathways of PC5B and furin are similar (De Bie et al. 1996).
PC7 is also transported on the constitutive secretory pathway via the TGN to the plasma membrane, from where it recycles via late endosomes to the TGN region, and concentrates SDV, a post-Golgi compartment distinguishable from the TGN (Wouters et al. 1998). PC7 shuttling between the plasma membrane and the TGN region depends on sequences in the cytoplasmic domain. The sorting signal for SDV localization consists of the following motifs: (1) peptide PLC726, (2) the basic amino acid sequence HRSRKAK708, and (3) two cysteines, C558 and C563, which are iteratively palmitoylated during the shuttle between TGN and plasma membrane (van de Loo et al. 2000; Declercq et al. 2012, 2017) (Fig. 9.5). Interestingly, a small fraction of PC7 reaches the cell surface through a brefeldin A and coat protein complex II (COPII)-independent unconventional secretory pathway. This may explain the rapid (<10 min) transit of PC7 from the ER to the cell surface (Rousselet et al. 2011), whereas the cleavage of the propeptide of PC7 is a slow process which takes hours rather than minutes (Creemers et al. 2000).
PC1/3 targeting to dense-core secretory granules (DCSG) resides in signals of the carboxy terminal ectodomain (617–753) which contains two α-helices, helix 1 (722–728) and helix 2 (738–750), of which the last one is sufficient for targeting a constitutively secreted protein to dense-core secretory granules (Dikeakos et al. 2009).
SKI-1/S1P cleaves cellular substrates in the ER/cis-Golgi area. There are differences, however, with arenaviral glycoproteins. The Lassa virus glycoprotein is cleaved by SKI-1/S1P before reaching the cis-Golgi, whereas cleavage of the LMCV glycoprotein occurs in the late or post-Golgi compartment (Lenz et al. 2001; Beyer et al. 2003). The substrate recognition of cleavage site variants is dependent on the auto-processing of SKI-1/S1P, suggesting differences in the processing of cellular and viral substrates (Burri et al. 2012a).
3 Proprotein Convertases Activating Viruses
Furin and the furin-like PCs, cleaving peptide bonds after basic residues, as well as SKI-1/S1P which cleaves peptide bonds after nonbasic residues, are the PCs which cleave viral glycoproteins (Table 9.3). Furin and furin-like PCs (PC5/6, PACE4, and PC7) are widely or ubiquitously expressed and are responsible for most of the processing events occurring in the constitutive secretory pathway or in endosomes. This leads to the activation/inactivation of receptors, ligands, enzymes, viral glycoproteins, or growth factors. Although these PCs exhibit a certain degree of functional redundancy when overexpressed in cell lines, their inactivation in mice or human beings results in specific phenotypes revealing that, in vivo, each PC primarily fulfills unique processing events and/or functions (Seidah et al. 2013). Involvement of other proteases with similar specificity cannot be excluded, but has not been demonstrated so far.
3.1 Furin
Furin , also named PACE or PCSK3, is the prototype of subtilisin-/kexin-like proprotein convertases (PCSKs). Furin is encoded by a transcription unit in the upstream region of the c-fes/fps proto-oncogene (Roebroek et al. 1986). Furin is expressed in all cells and tissues of eukaryotic organisms. It is synthesized as pro-furin with a molecular mass of 100 kDa, which is autocatalytically cleaved into the mature form with a molecular mass of 85 kDa. The first cleavage occurs between the prodomain and the catalytic domain at the C-terminus of the motif 101AKRRAKR↓ and the second one within the prodomain at the amino acid motif 70 RGVTKR↓ (Leduc et al. 1992; Anderson et al. 1997). Endoproteolytic cleavage and removal of the propeptide fragments are prerequisite for efficient transport out of the endoplasmic reticulum into the TGN where furin acquires full enzymatic activity (Creemers et al. 1995). Endogenous furin was partially purified from Madin-Darby bovine kidney (MDBK) cells and identified by reaction with a furin-specific antiserum (Stieneke-Gröber et al. 1992; Vey et al. 1994). Furin is partially cleaved at arginine (R683) present in the ectodomain by an unknown endoprotease residing at the plasma membrane. The truncated soluble furin is catalytically active outside of cells (Plaimauer et al. 2001).
Furin is the central proprotein convertase that processes most diverse proproteins at multibasic structures on the constitutive secretory pathway. An analysis of the human proteome revealed an estimated number of about 500 potential proprotein candidates susceptible to furin cleavage (Remacle et al. 2008; Shiryaev et al. 2013). The high number of potential substrates together with the ubiquitous expression implicates that furin activates a wide variety of membrane-anchored proteins and membrane-secreted proteins, including precursors of growth factors, cell receptors, adhesion molecules, matrix metalloproteinases, blood plasma proteins, and factors for embryonal development which play important roles in the regulation of many life processes (Table 9.2). Furin also generates MHC class I antigens (Gil-Torregrosa et al. 2000). The majority of the proproteins are cleaved at the multibasic motif RXK/RR and less frequently at the minimal basic motif RXXR. A few precursor proteins possess exceptional motifs, e.g., consensus sequences without arginine or lysine at position P4 as observed with the prodomain of furin (RGVTKR), proalbumin (RGVFRR), proprotein C (RSHLKR), and proparathyroid hormone (KSVKKR) (Canaff et al. 1999; Mori et al. 1999; Essalmani et al. 2017). Single amino acid positions of the furin motif were extensively studied (Rockwell et al. 2002). Proteins with lysine at position P4 are poor furin substrates but are readily cleaved by TMPRSS13 belonging to the family of transmembrane serine proteases (cf. Chap. 8).
The fact that the hemagglutinin precursor (HA0) of FPV containing a multibasic cleavage site was correctly cleaved and is biologically active after expression in insect cells and in insect larvae indicated that endogenous furin exists in insects (Kuroda et al. 1986, 1989). Furin of Spodoptera frugiperda showed the same cleavage properties as furin of vertebrates (Cieplik et al. 1998). In Drosophila melanogaster, two genes homologous to human furin, called Dfur1 and Dfur2, have been identified. The Dfur1 gene undergoes differential splicing to generate several type I membrane-bound isoenzymes differing in their C-terminal sequences. They are released as soluble dfurin forms which show cleavage specificity like furin of mammalian species (De Bie et al. 1995; Roebroek et al. 1991).
Furin is expressed in the mouse embryo at embryonic day e7.5. Inactivation of the fur locus by homologous recombination in the mouse causes embryonic death shortly after e10.5 due to hemodynamic insufficiency and failure of ventral closure and axial rotation in embryos. The furin-deficient mouse embryos failed to develop large vessels despite the presence of endothelial cell precursors (Roebroek et al. 1998). Among numerous proteins which play crucial roles during embryonic development are transforming growth factor β1 (TGFβ1), bone morphogenetic proteins 5 and 7 (BMP5, BMP7), vascular cell adhesion molecule (VCAM-1), and α-integrins (Scamuffa et al. 2006). To overcome the lethality of furin knockout mice, conditional knockout mutants were constructed. When furin expression was switched off in an interferon-inducible Mx-Cre/loxP knockout mouse model, the animals showed no obvious adverse effects. Histological analysis of the liver did not reveal any overt deviations from normal morphology. Variable degrees of redundancy were observed for the processing of numerous substrates, but none of the tested substrates displayed a complete block of processing. The absence of a severe phenotype raises the possibility of using furin as a local therapeutic target in the treatment of pathologies like cancer and viral infections, although the observed redundancy may require combination therapy or the development of a more broad-spectrum convertase inhibitor (Roebroek et al. 2004; Creemers and Khatib 2008).
Furin plays an important role in virus activation . As mentioned above, the first viral protein found to be processed by furin was the hemagglutinin (subtype H7) of fowl plague virus (FPV) (Stieneke-Gröber et al. 1992). Studies on the hemagglutinin of FPV and viruses of subtype H5 had indicated the presence of a multibasic cleavage site (Bosch et al. 1981; Kawaoka et al. 1987; Kawaoka and Webster 1988), and mutational analyses of the H7 cleavage site clearly defined the characteristic RXK/RR motif. Cleavage occurs only when this motif is presented in the correct sequence position in loop formation. A shift of the motif by only one amino acid can abrogate cleavage (Garten et al. 1991; Vey et al. 1992). The importance of conserved amino acids of the hemagglutinin of HPAIV was corroborated, especially an arginine at P1 position proved to be essential (Walker and Kawaoka 1993; Walker et al. 1994). The hemagglutinin cleavability of HPAIV is influenced by the amino acid immediately downstream of the cleavage site (Horimoto and Kawaoka 1995). Similar results were obtained for the cleavage motifs of the highly pathogenic Newcastle disease virus (NDV) strains (Pritzer et al. 1990; Gotoh et al. 1992).
Over nearly three decades of research, an increasing number of fusion-competent glycoproteins have been identified which are activated by furin or furin-like proteases. Most glycoproteins belong to the enveloped RNA viruses, but also some enveloped DNA viruses use furin cleavage for maturation (Table 9.3). Accessory proteins forming a complex with fusion proteins, such as the prM protein of flaviviruses, are also activated by furin (cf. Chap. 6). However, furin can be replaced by another furin-like protease in certain cell types and tissues. Such examples were observed with HPAIV and with HIV-1 (Feldmann et al. 2000; Horimoto et al. 1994; Hallenberger et al. 1992; Anderson et al. 1993; Gu et al. 1995; Ohnishi et al. 1994). The glycoproteins of influenza viruses and paramyxoviruses are activated by furin next to the fusion peptide. Other viral glycoproteins are cleaved by furin at more than one site, as is the case with respiratory syncytial virus (RSV) (see below), or they are cleaved by furin in concert with other endoproteases as found with coronaviruses (cf. Chap. 4).
Detailed lists of more furin-cleavable viral membrane proteins are given in Table 9.3 and in a previous review (Klenk and Garten 1994). All of these proteins have multibasic cleavage sites, but experimental evidence for furin cleavage has not been obtained in all cases.
Various inhibitors blocking the catalytic activity have been used to substantiate the role of furin as processing enzyme. The first inhibitory agents were acylated basic tetrapeptidyl chloromethyl ketones , such as Dec-RVKR-CMK, polyarginines (nona-d-arginine amide), and serpin inhibitors , such as furin-adapted α-1-antitrypsin Portland (α-1-PDX) (Garten et al. 1989, 1994; Misumi et al. 1990a; Molloy et al. 1992; Jean et al. 1998; Cameron et al. 2000; Kacprzak et al. 2004; Hardes et al. 2015, 2017) (cf. Chap. 11). Acylated peptidyl chloromethyl ketones containing the RXK/R R motif bind covalently to the catalytic site of furin and prevent cleavage activation of HPAIV hemagglutinin and a wide array of other fusion-competent viral glycoproteins (Garten et al. 1994). In early years, when furin was still the only PC known substrate, homologous inhibitors were thought to be useful for the identification of furin as the activating protease of a virus. However, this perspective changed when other PCs were discovered. Because many of these enzymes have similar substrate specificities, the inhibitors were of limited use to discriminate them from furin. More convincing approaches for discrimination of PCs with closely related substrate specificity are investigations which exploit cells or animals with selective deficiency of a single PC or approaches which knock down the mRNA of a distinct PC.
3.2 PACE4
Paired basic amino acid-cleaving enzyme 4 (PACE4) , also known as subtilisin-/kexin-like proprotein convertase (PCSK6), exists as alternatively spliced transcript variants encoding various isoforms which are differently present in tissues and cells. PACE4 is mainly expressed in liver, spleen, gut, brain, and neuroendocrine cells (Seidah et al. 2008). PACE4 migrates on the constitutive pathway and is secreted into the extracellular matrix, where it is attached to heparan sulfate proteoglycans (HSPGs) (Tsuji et al. 2003). PACE4 shares many substrates with furin, such as TGFβ-related proteins, proalbumin, pro-von Willebrand factor, zymogens of the a disintegrin and metalloprotease with thrombospondin type I motif (ADAMT) family, and the precursor of the low-density lipoprotein receptor (pro-LDL receptor). However, it has a more stringent substrate specificity and more limited operating parameters than furin (Longpré and Leduc 2004; Seidah et al. 2013; Wong et al. 2015) (Table 9.2). PACE4 processes diphtheria toxin and anthrax toxin protective antigen, but not Pseudomonas exotoxin A which is processed by furin (Moehring et al. 1993; Sucic et al. 1999). PACE4 cleaves at RXK/RR, RXXR and to a much lesser extent at RR and KR motifs (Gordon et al. 1997). In contrast to furin, PACE4 cannot process pro-factor IX and is not inhibited by the alpha1-antitrypsin Portland variant (Rehemtulla et al. 1993; Mains et al. 1997). In the absence of PACE4, mouse embryos developed specific deficiencies and survived to 75% compared with 100% lethality of furin knockout mice (Table 9.1). This suggests that, although PACE4 and furin share the ability to process similar substrates, they may also process one or more different substrates during the processes of embryonic development (Scamuffa et al. 2006). There are only few studies demonstrating cleavage of viral glycoproteins by PACE4 (Table 9.3).
3.3 PC5
PC5 is identical with PC6 and often designated PC5/6 or PCSK5 . The proprotein convertase 5 gene is transcribed into two mRNAs; consequently two different but related enzymes are formed: PC5A and PC5B comprising 913 and 1860 amino acids, respectively. The A and B isoforms have identical pro-, catalytic, and P-domains. The catalytic domain (amino acids 148–439) contains D171, H212, N313, and S386 in the active-site pocket. Both isoforms differ in the cysteine-rich domain (CRD) (Fig. 9.1). PC5A has a CRD domain in comparable length with other PCs and terminates in the ectodomain as a soluble enzyme. PC5A migrates on the regulated secretory route and is packaged into dense-core granules. In contrast, PC5B, containing besides an unusually long CRD domain a transmembrane and a cytoplasmic domain, is transported on the constitutive branch of the secretory pathway. PC5B can be shed from the membrane, and the soluble PC5A can be attached to the cell surface by interaction of its CRD with proteoglycans. A PC5B transcript was found mainly in the intestine and kidney, while PC5A transcripts were detected in various tissues indicating different locations and roles for PC5A and PC5B (Nakagawa et al. 1993). PC5A is the major isoform in most tissues analyzed, except in the liver, where the transcripts are expressed in equivalent amounts. The complete knockout of PC5 in mice causes death at birth, with the embryos exhibiting multiple morphogenic defects (Table 9.1). Conditional knockout mice revealed that growth differentiation factor 11 (Gdf11), also known as bone morphogenetic protein 11 (BMP-11) and BMP-2, is a favorite substrate of PC5; other substrate precursor proteins are vascular endothelial growth factors (Essalmani et al. 2008; Lee et al. 2015). PC5 plays also a dominant role in pregnancy establishment by proteolytic activation of several important factors such as BMP2, caldesmon 1, calmodulin- and actin-binding protein (CALD1), and α-integrins.
The deduced cDNA structures of mouse PC5 and rat PC5 showed that the closest homologue is PACE4. Furthermore, like furin, Drosophila melanogaster dfurin2, and PACE4, PC5 shows the presence of a C-terminal cysteine-rich domain containing either five (PC5 and PACE4) or ten (dfurin2) repeats of the consensus motif Cys-Xaa2-Cys-Xaa3-Cys-Xaa (5-7)-Cys-Xaa2-Cys-Xaa (8-15)-Cys-Xaa3-Cys-Xaa (9-16). The richest sources of rat PC5 mRNA (3.8 kb) are the adrenal gland and gut, but it can also be detected in many other endocrine and nonendocrine tissues (Lusson et al. 1993).
PC5 activates the hemagglutinin of HPAIV like furin (Horimoto et al. 1994; Feldmann et al. 2000) (Table 9.3).
The decapeptide 107QQVVKKRTKR 116 mimicking a part of the prodomain of proPC5 is a nanomolar inhibitor of furin, PACE4, and PC5. A mutation at position P6 (K111H) makes the inhibitor more selective for PC5 than for furin indicating that a modification around the basic motif may influence the selectivity of PCs (Nour et al. 2003).
3.4 PC7
PC7 is identical with PC8, PCSK7, and lymphoma proprotein convertase (LPC) as it was originally discovered in a high-grade lymphoma carrying a translocation (Meerabux et al. 1996). PC7 is the most ancient and conserved member of the PC family. It is synthesized as proenzyme (101 kDa) and autocatalytically processed into mature PC7 (89 kDa) at RRAKR 141↓. There is no truncated soluble form of PC7 (Declercq et al. 2017). PC7 is invariably expressed as a membrane-anchored enzyme in spleen, thymus, prostate, testis, ovary, small intestine, colon, and peripheral blood leukocytes. High levels of PC7 mRNA are found in cells of the immune system, particularly in CD8+ cells, but also in CD4+, NK cells, and bone marrow cells. This may indicate a role in immune functions. The first described PC7-specific processing reaction was the activation of epidermal growth factor receptor (EGFR) at the cell surface (Rousselet et al. 2011). PC7 cleaves the unusual peptide KSVKKR↓SVSEIQL derived from proparathyroid hormone. Coexpression of PC7 and human transferrin receptor 1 (hTfR1) indicated that PC7 is the only convertase that sheds this receptor from cells into the medium, whereby the cleavage occurs at the site 95 KTECER↓ LA resembling the cleavage site of parathyroid hormone (Guillemot et al. 2013). The cleavage specificity of PC7 is largely similar to that of furin with the motifs (K/R)R↓ or (R/K)XnR↓ (n = 2, 4, or 6 amino acids). A 24-mer peptide fragment of the PC7 prosegment (residues 81–104) is a strong inhibitor, K(i) = 7 nM of PC7, comparable to that of the full-length (104 residue) prosegment (Bhattacharjya et al. 2000). Unlike other PC-deficient mice, PC7-null mouse embryos did not show an apparent abnormal phenotype supporting the view that PC7 expression extensively overlaps with that of furin (Seidah 2011). However, PC7 is essential for zebrafish development and bioavailability of TGFβ1a. When the PCSK7 function in developing larvae was inhibited, defects in various organs including the brain and eye were observed, and the larvae died within 7 days postfertilization (Turpeinen et al. 2013). Expression of PC7 revealed an increased ADAM10 maturation resulting in enhanced α-secretase-mediated processing of amyloid precursor protein (Anders et al. 2001; Lopez-Perez et al. 2001). PC7 cleaves specifically and in a cell-type-specific manner gp160 of HIV into gp120g/p41, suggesting that both furin and PC7 are the major convertases in T4 lymphocytes (Decroly et al. 1997; Hallenberger et al. 1997).
In summary, the furin-like PCs PACE4, PC5, and PC7 may selectively compensate the activation of viral glycoproteins in furin-deficient cells and tissues with different efficiencies. Furin compensation has been demonstrated (1) for virulent NDV F protein activated by PC5, but not by PACE4 (Fujii et al. 1999); (2) for processing of E3/E2 from Chikungunya virus by PC5A, PC5B, and PACE4, but not by PC7 (Ozden et al. 2008); (3) for cleavage of peptides homologous to SARS coronavirus S glycoprotein by PC5, but not by PC7 (Basak et al. 2007); and (4) for activating the env of HIV by PC7 (Decroly et al. 1997; Hallenberger et al. 1997).
3.5 SKI-1/S1P
This enzyme is known under the names subtilisin-/kexin-isozyme-1 (SKI-1) , site 1 protease (S1P) membrane-bound transcription factor peptidase site 1, sterol regulatory element-binding protein site 1 (SREBP S1) protease, membrane-bound transcription factor protease site 1 (MBTPS1), and PCSK8. The protease plays an important role in lipid metabolism. The lipid composition of animal cells is controlled by SREBPs, transcription factors released from membranes by sterol-regulated proteolysis. By comparing cDNA of protease-competent and protease-deficient cells, the group of Goldstein and Brown identified the enzyme, which they called S1P, as an intraluminal membrane-bound subtilisin-like protease, 1052 amino acid long, which cleaves SRBPs at the motif RSVL↓ in the ER luminal loop between two membrane-spanning regions (Sakai et al. 1998). Using reverse transcriptase (RT)-PCR and degenerated oligonucleotides derived from the active-site residues of subtilisin-/kexin-like serine proteinases, Seidah and colleagues independently identified a highly conserved and phylogenetically ancestral human, rat, and mouse type I membrane-bound proteinase which they called subtilisin-/kexin-isozyme-1 (SKI-1) (Seidah et al. 1999). The tissue distribution of SKI-1/S1-P mRNA is ubiquitous. SKI-1/S1P accumulates in the perinuclear region, predominantly in the cis-Golgi, and small punctual SKI-1/S1P containing material is seen in the endosomal/lysosomal compartments by immunochemical staining. Studies with brefeldin A indicated that substrates are cleaved in the early Golgi. proSKI-1/S1P is processed into two membrane-bound forms of SKI-1 (120 and 106 kDa) differing by the nature of their N-glycosylation (da Palma et al. 2014, 2016). At late stages of the secretory pathway, part of the membrane-bound enzyme is shed into the medium in a 98-kDa form. SKI-1/S1P exhibits a wide pH optimum for cleavage (Seidah et al. 1999). Recombinant SKI-1/S1P was expressed, purified, and characterized (Bodvard et al. 2007).
A physiological SKI-1/S1P substrate different from SREBPs is ATF6, a membrane-bound transcription factor that activates genes in ER stress, such as cholesterol deprivation. When unfolded proteins accumulate in response to ER stress, ATF6 is cleaved at RHLL↓ to release its cytoplasmic domain, which enters the nucleus (Ye et al. 2000). Other substrates are (pro)renin receptor and brain-derived neurotrophic factor (BDNF) (Nakagawa et al. 2017). BDNF is a member of the neurotrophin family of growth factors found in the brain and the periphery which is cleaved at RGLT↓. SKI-1/S1P is required for the transcription of many bone matrix and mineralization-related genes, such as fibronectin and fibrillin in bone osteoblasts and osteocytes. The irreversible inhibitor Dec-Arg-Arg-Leu-Leu-CMK blocks transcription of the corresponding genes and inhibits mineralization (Gorski et al. 2011). These results demonstrated that the differentiated phenotype of osteoblastic cells and possibly osteocytes depends upon SKI-1/S1P. Knockdown of SKI-1/S1P in zebrafish leads to the zebrafish gonzo mutant showing a defect in chondrocyte morphogenesis (Schlombs et al. 2003).
Since SKI-1/S1P plays an essential role in cell physiology, it is evident that gene deletion is lethal at an early state of the embryonal development (Table 9.1). Conditional SKI-1/S1P knockout mice are viable (Yang et al. 2001; Seidah 2011).
The SKI-1/S1P processing motifs contain basic and hydrophobic residues at P4 and P2, respectively, with a relatively relaxed acceptance of amino acids at P1 and P3, i.e., R4-X3-X2-X1 ↓, where X1,3 are any amino acids and X2 is often leucine, isoleucine, and valine. A favorable motif is RRLL↓ found in the glycoprotein of Lassa virus (Maisa et al. 2009). The first viral proteins recognized to be cleaved by SKI-1/S1P were the glycoproteins of Lassa virus and lymphocytic choriomeningitis virus (LCMV) (Lenz et al. 2000, 2001; Beyer et al. 2003; Kunz et al. 2003). A high variability of cleavage motifs is found with glycoproteins of arenaviruses and Crimean-Congo hemorrhagic fever virus (CCHFV) (Burri et al. 2012b, 2013; Altamura et al. 2007; Sanchez et al. 2002, 2006; Vincent et al. 2003) (cf. Chap. 3). SKI-1/S1P inhibition effectively blocks hepatitis C virus (HCV) from establishing infection in hepatoma cells (Olmstead et al. 2012). However, the function of SKI-1/S1P in HCV replication is not known.
4 PCs Not Known to Activate Viruses
4.1 PC1/3
PC1/3 , also designated neuroendocrine convertase 1 (NEC1), prohormone convertase 3 (PC3), proprotein convertase 1 (PC1), or PCSK1, occurs in many eukaryotic organisms. The human gene of PC1/3 located on chromosome 5 is transcribed from 13 exons and translated into the preproprotein of PC1/3 comprising 753 amino acids. The catalytic domain (143–441 aa) contains the triad D167, H208, and S382. N309 contributes to the oxyanion hole and stabilizes the transition state of the enzymatic reaction. The catalytic domain has 61% sequence homology with human furin. The prodomain contains 83 amino acids with 30 to 40% sequence identity among the eukaryotic PC1/3s. PC1/3 is present in dense-core vesicles of the regulated secretory pathway in neuroendocrine tissues. It cleaves prohormones and other precursor proteins C-terminally at arginine-arginine or lysine-arginine motifs (Table 9.1). Typical substrates are POMC, proinsulin, proglucagon, and other precursors of neuro-sensing and regulating hormone peptides, such as the hormone-like endopeptidase renin, enkephalin, dynorphin, somatostatin, ghrelin, and agouti-related protein (AGRP) (Creemers et al. 2006). Interestingly, PC1/3 deficiency has quite different phenotypic effects in mice and man, a phenomenon not observed with other PCs. PC1/3 knockout mice are viable but exhibit growth retardation and multiple defects in hormone precursor processing. In humans, PC1/3 deficiency causes obesity, hypogonadism, reactive hypoglycemia, hypoadrenalism, and small-intestinal absorptive dysfunction due to impaired processing of prohormones (Taylor et al. 2003; Farooqi et al. 2007; Seidah 2011) (Table 9.1).
4.2 PC2
PC2 is also known as neuroendocrine convertase 2 , Kex-like endoprotease 2, or PCSK2. Maturation of PC2 is unusual since it depends on support by the neuroendocrine chaperone 7B2 that prevents auto-aggregation of PC2 (Ramos-Molina and Lindberg 2015). After binding of the chaperone to nascent proPC2, the proPC2/7B2 complex is transported from the ER to the TGN. On the route from the TGN to dense secretory granules, the chaperone dissociates from the complex in the acidic environment, and the prodomain is removed after autocatalytic cleavage (Seidah 2011). Embedding of the C-terminal domain of PC2, and likewise of PC5A, in glycosphingolipid- and cholesterol-rich microdomains appears to be necessary for sorting into secretory granules (Creemers et al. 1996; De Bie et al. 1996). Evidence has also been obtained that an amphipathic α-helix at the C-terminus serves to fix PC5A at the cell membrane (Assadi et al. 2004). Like PC1/3, PC2 cleaves at dibasic amino acid motifs of neuroendocrine peptide and protein precursors, such as pro-opiomelanocortin (POMC), proinsulin, and proglucagon, and the precursors of chromogranin A, neurotensin, and pro-enkephalin (Pan et al. 2006).
PC2 −/− mice appear normal at birth but show retarded growth with chronic fasting, hypoglycemia, and reduced glucagon levels (Seidah 2011).
4.3 PC4
PC4 , also known as proprotein convertase subtilisin/kexin type 4 (PCSK4) , is expressed from 15 exons which code for the mRNA of a polypeptide containing 655 amino acids. PC4 is found exclusively in germ cells, suggesting a possible reproductive function of this enzyme (Basak et al. 1999; Nakayama et al. 1992; Seidah et al. 1992). The catalytic domain with its enzymatic triad D158, H189, S373 and the oxyanion hole-forming N300 processes various proproteins by cleavage at paired basic amino acids of the general motif (R,K)X(R,K,X)R. PC4 is closely related to furin. It is primarily found in testicular germ cells and in sperm but also in ovary macrophages. It controls testicular and ovarian physiology.
PC4 is expressed in the human placenta and cleaves the precursor of the insulin-like growth factor II, an important regulator of fetoplacental growth (Qiu et al. 2005). Another specific substrate of PC4 in the testis is pituitary adenylate cyclase-activating polypeptide (PACAP) which is solely processed by PC4 (Seidah 2011). The fertility of PC4 −/− mice is significantly reduced (Scamuffa et al. 2006), and there are also defects in embryonic development (Mbikay et al. 1997).
4.4 PCSK9
PCSK9 , also called neural apoptosis-regulated convertase-1 (NARC-1) , is a new member of the pyrolysin and proteinase K subfamilies of subtilases (Abifadel et al. 2003). It is highly expressed in the liver as a 74 kDa protein that is autocatalytically cleaved. The prodomain remains tightly bound to the enzyme which is catalytically inactive but functions as a binding protein that interacts with the low-density lipoprotein receptor (LDLR) and plays therefore a crucial role in plasma cholesterol homeostasis. PSCK9 is not involved in proteolytic protein processing.
Loss-of-function PCSK9 mutations were also identified. Two nonsense mutations found in ∼2% of black Africans were associated with a ∼40% decrease in LDL cholesterol and a ∼88% risk reduction of cardiovascular disease (CVD), suggesting that PCSK9 inhibition may be a promising approach to treat hypercholesterolemia and prevent CVD. Therapeutic monoclonal antibodies have been developed to reduce low-density lipoprotein (LDL) cholesterol levels and the risk of coronary artery disease (Weider et al. 2016; Le et al. 2015; Seidah et al. 2017).
5 Functions of PCs in Virus Replication
5.1 Cleavage Activation of Viral Fusion Proteins
Enveloped viruses induce fusion of viral and cellular membranes to deliver their genomes into the cytoplasm. All viral fusion proteins are C-terminally anchored in the viral membrane and possess a hydrophobic “fusion peptide” (class I fusion proteins) or “fusion loop” (class II and class III fusion proteins) which interacts with the target membrane. Exposure of the hydrophobic domains depends on a conformational change of the fusion protein triggered by low pH in endosomes or by interaction with the receptor-binding protein at the cell surface (Harrison 2008; White and Whittaker 2016; Jardetzky and Lamb 2014). In most cases, the conformational change can only be triggered if the proteins are primed by proteolytic cleavage.
For many viruses, like influenza viruses, paramyxoviruses, retroviruses, and arenaviruses, the cleavage site is a distinct peptide bond next to the “fusion peptide” which, after cleavage, leads to a new exposed N-terminus of the membrane-anchored fusogenic subunit of the viral glycoproteins. The fusion peptide is characterized by a stretch of non-charged hydrophobic amino acids which are strongly conserved within a virus family. In contrast to the conserved fusion peptides, the preceding cleavage sites and the activating proteases vary even when the viruses are closely related. Multibasic cleavage sites recognized by furin and furin-like proprotein convertases PACE4, PC5/6, and PC7 are present in most enveloped viruses, whereby furin is the master key for cleavage of viral glycoproteins.
Furin is also involved in a different fusion mechanism observed with flaviviruses including tick-borne encephalitis virus (TBEV), yellow fever virus, West Nile virus, dengue virus, and Zika virus. Envelope glycoprotein E of these viruses, a class II fusion protein, is not processed by proteolysis. Fusion activity depends, however, on furin cleavage of the tightly associated accessory protein prM. During intracellular virion assembly, prM prevents premature exposure of the fusion loop located on E. After cleavage of prM and release of the prepeptide, mature virions invade cells by endocytosis and exposure of the fusion loop at low pH (cf. Chap. 6). Blockage of prM cleavage by furin inhibitors prevents multiple replication cycles of TBEV and dengue virus in cell cultures (Stadler et al. 1997; Elshuber et al. 2003; Kouretova et al. 2017).
Enveloped DNA viruses utilize the fusion mechanism for host cell invasion, too. Membrane fusion during herpesvirus entry into host cells is a complex process. All herpesviruses express the gB and gHgL complex as well as various non-conserved glycoproteins of individual herpesviruses which interact with selected receptor proteins determining cell tropism (Eisenberg et al. 2012). The gHgL complex acts as an “activator” of entry, and the glycoprotein gB acts as the membrane “fusogen.” It has been shown that gB of several herpes viruses is cleaved by furin (Table 9.3). gB is a class III fusion protein with a “fusion loop” and has structural domain similarities with VSV G protein and the baculovirus fusion protein. The cleavage of Epstein-Barr virus gB is required for cell-cell fusion (Sorem and Longnecker 2009), whereas bovine herpesvirus 1 gB and pseudorabies virus gB are not necessarily cleaved (Kopp et al. 1994). Baculovirus nucleopolyhedrovirus (NPV) group II members have a fusion protein, which must be cleaved by furin to mediate fusion (Long et al. 2006; Wang et al. 2017).
5.2 Cooperation of Proprotein Convertases with Other Endoproteases
Virus surface glycoproteins of several viruses are activated by various endoproteases at different sites. The spike protein S of coronaviruses which possesses both receptor-binding and fusion functions, is cleaved at several sites (Table 9.3). For example, the severe acute respiratory syndrome (SARS) coronavirus glycoprotein S is cleaved at two sites, 441 RYLR↓ and 758 RNTR↓ (S1/S2 site), by furin/furin-like proteases, and at the S2´cleavage site 796 KR↓ by cathepsin L or transmembrane serine protease 2 (TMPRSS2). The proteolytic activation of protein S at amino acid position 797 is adjacent to the “fusion peptide” of the S protein, which proved to be crucial for fusogenicity. Similarly, the Middle East respiratory syndrome (MERS) coronavirus possesses a glycoprotein S that is cleavable at three distinct sites by furin (Table 9.3). Modulations of the spike cleavage S2′ next to the fusion peptide have profound effects on tropism and pathogenicity (Belouzard et al. 2009; Millet and Whittaker 2015; cf. Chap. 4).
Glycoprotein GP of Ebola virus mediating receptor binding and fusion is first processed to GP1/2 at a conserved furin cleavage site 497RRTRR↓ which is remote from the fusion loop located on GP2 (amino acids 524 to 540) (Volchkov et al. 1998, 2000). Furin processing is followed by cathepsin cleavage at about amino acid 200 resulting in the removal of a large C-terminal fragment of GP1 and the exposure of a receptor-binding site (Chandran et al. 2005). Both processing steps are therefore essential for the function of GP in virus entry. There is evidence that furin and cathepsin can be replaced in GP processing by other proteases. Furin cleavage of a nonstructural glycoprotein (sGP) of Ebola virus has also been observed (cf. Chap. 5).
The genome segment M of Crimean-Congo hemorrhagic fever virus (CCHFV) polyprotein encodes a polyprotein with four transmembrane anchors separating three luminal/extracellular domains from two cytoplasmic domains. Furin and SKI-1/S1P are involved in the activation mechanism of this polyprotein (Sanchez et al. 2002, 2006; Altamura et al. 2007). Cleavages by SKI-1/S1P and by furin are necessary for producing the nonstructural glycoprotein GP38 and the structural glycoprotein Gn, an important step for gaining fusion capacity (Bergeron et al. 2015).
The envelope glycoprotein (env) of foamy virus, a spuma retrovirus, shows an unusual biosynthesis undergoing cleavage by furin at two different positions. The precursor protein has a type III membrane topology with both the N and C termini located in the cytoplasm. The processing of env into the particle-associated env leader protein (elp) and into the surface (SU) and transmembrane (TM) subunits occurs posttranslationally during transport to the cell surface. Furin or a furin-like protease is responsible for both, the late signal peptidase-like processing and the maturation cleavage needed for fusion (Duda et al. 2004; Geiselhart et al. 2004).
5.3 Generation of Biologically Active Peptides
The fusion protein of the human and bovine respiratory syncytial viruses (RSV) is synthesized as an inactive precursor F0 that is proteolytically processed at two multibasic sequences, 131KKRKRR↓ and 106 RARR↓ (bovine RSV). Both furin consensus sequences must be cleaved to activate the fusion protein (Zimmer et al. 2001; González-Reyes et al. 2001). Cleavage of the bovine RSV fusion protein results in the release of a small peptide that is converted into the biologically active virokinin by additional posttranslational C-terminal modifications, namely, truncation by carboxypeptidase of B type and stabilization by enzymatic amidation. The amino acids 106–139 of the viral fusion protein align with amino acids 45–78 of the human tachykinin precursor type 1 that functions as a tissue hormone and produces rapid contraction of smooth muscles. Virokinin is secreted by virus-infected cells and was found to desensitize tachykinin receptors in the mammalian respiratory tract with potent effects on local inflammatory and immune processes (Zimmer et al. 2003).
5.4 Glycoprotein Trimerization and Incorporation into Virus Particles
Cleavage of the Lassa virus glycoprotein by SKI-1/S1P is necessary not only for fusion of the virus envelope with endosomal membranes at cell entry but also for efficient incorporation of the viral glycoprotein into virus particles (Lenz et al. 2001). Virions contain almost exclusively homotrimeric spikes of the cleaved glycoprotein form, whereas at the cell surface of infected cells, monomers and oligomers of the uncleaved form prevail (Schlie et al. 2010a). Glycosylation mutations on the ectodomains of Lassa virus glycoprotein showed that 6 of 11 N-glycans are necessary for glycoprotein cleavage indicating that N-glycans are needed for correct conformation of the precursor glycoprotein to be cleaved by SKI-1/S1P. Interestingly, the glycoprotein precursor is transported to the cell surface in a completely endo H-sensitive form suggesting that cleavage is a prerequisite for completion of complex N-glycosylation (Eichler et al. 2006). Moreover, a mutation in the cytoplasmic domain of Lassa virus glycoprotein abolished the maturation cleavage by SKI-1/S1P within the ectodomain indicating conformational changes across the membrane (Schlie et al. 2010b). These data suggest that, with some viruses, cleavage by PCs may be important for correct folding, oligomerization, and N-glycosylation, which is assumed to be a prerequisite for an effective incorporation of functional virus spikes into virions. Extensive structural rearrangements in the Lassa surface glycoprotein were observed at different pH’s and in the presence of the functional secondary intracellular receptor, human LAMP-1, by using high-resolution electron cryo-microscopy and tomography techniques (Li et al. 2016). Such a substantial rearrangement of subdomains of the Lassa viral glycoproteins would not be possible in the absence of SKI-1/S1P cleavage.
The glycoproteins of Borna disease virus (BDV) and simian immunodeficiency virus (SIV ) also need to be cleaved by furin for correct trimerization and effective insertion into virus particles (Eickmann et al. 2005; Yamshchikov et al. 1995).
5.5 PC Cleavage of Non-Envelope Proteins
Viral protein R (Vpr) is a HIV-1 protein 96 amino acid long that plays an important role in regulating nuclear import of the HIV-1 pre-integration complex and is required for virus replication in nondividing cells such as macrophages. It has been detected in soluble form in the sera and cerebrospinal fluids of HIV-1-infected patients and contributes to HIV pathogenesis. Vpr was found to undergo proteolytic processing at the PC cleavage site, 85 RQRR↓ located within the functionally important C-terminal arginine-rich domain. Interestingly, Vpr processing occurred extracellularly upon close contact to cells and most likely involved cell surface-associated furin or furin-like PC (Xiao et al. 2008).
Hepatitis B virus e-antigen (HBeAg) is a secreted version of hepatitis B virus (HBV) core protein raised from the core gene. HBeAg has a cleavable signal peptide and four furin cleavage sites in the C-terminal part. HBeAg promotes immune tolerance and is essential for the development of chronic hepatitis B virus infection. Furin plays a key role in processing the HBeAg precursor into mature HBeAg (Ito et al. 2009; Pang et al. 2013). The furin inhibitor decanoyl-RVKR-chloromethylketone (CMK) combined with the nucleoside analogue entecavir reduced HBV replication and HBeAg secretion in Hep cells and was suggested as a therapeutic regimen for treatment of chronic hepatitis B (Yang et al. 2014).
Papillomaviruses (PV) are non-enveloped viruses that enter cells by endocytosis. Entry involves removal of protein L2 from the surface of the virus particles by furin (Day et al. 2008; Richards et al. 2006).
5.6 Protease Activation Mutants for Vaccine Design
A large number of the viruses activated by furin which are shown in Table 9.3 cause important diseases. Highly pathogenic avian H5 and H7 influenza viruses (HPAIV) are not only a problem for the poultry industry but are also a threat to human health. In 1997, an H5N1 virus caused human infections in Hong Kong during a poultry outbreak. Six of eighteen patients succumbed to this virus. Since 2003 the H5N1 virus spread rapidly killing millions of birds and continued to be transmitted to humans. Thus, there is a need for the development of human and animal vaccines against these viruses. For safety reasons the viruses used for the production of such vaccines should contain an HA that is modified at the furin cleavage site to diminish their pathogenic potential. This approach is recommended to meet short-termed demand of H5 or H7 vaccines (Neumann et al. 2008; Webby et al. 2004), and a vaccine with a monobasic HA cleavage site was licensed for humans in responsiveness to the pandemic alert in 2003 (Webby et al. 2004).
When an HA with a monobasic cleavage site is present in live vaccines, virus revertants with a furin cleavage site may emerge by spontaneous mutation. Therefore, safer vaccines are demanded that are produced from attenuated viruses containing a nonbasic cleavage site (Böttcher-Friebertshäuser et al. 2014). This concept is based on insights from early investigations in which attenuated mutants of Sendai and influenza viruses activated by elastase or chymotrypsin have been analyzed (Scheid and Choppin 1976; Orlich et al. 1995). More recently, this strategy was pursued to exchange the monobasic HA cleavage site of influenza strain A/WSN/33 (Stech et al. 2005) and the multibasic HA cleavage motif of an H7N7 influenza virus (Gabriel et al. 2008) for amino acids susceptible to elastase cleavage. The mutants were strictly elastase-dependent, grew equally well as the wild type in cell culture, and were attenuated in mice unlike the lethal wild type. Immunization with an H7N7 mutant at 106 pfu dosage protected mice against disease and induced sterile immunity; vaccination with homosubtypic or heterosubtypic reassortants led to cross protection. These observations demonstrate that a mutated HA requiring elastase cleavage can serve as an attenuating component of a safe live vaccine against HPAIV and other influenza viruses.
Flaviviruses with an altered furin cleavage site of prM have been shown to produce single-round infectious particles (Elshuber et al. 2003). Thus, it appears that protease activation mutants may also be a useful approach for the development of safe vaccines against other viruses.
5.7 Inhibitors of PCs for Host-Directed Antiviral Therapy
The high dependence of many viruses on PCs makes PCs promising targets for antiviral therapy. Furin inhibitors comprising substrate homologous peptidomimetics, furin- or SKI-1/SP1-adapted serpines, and blocking antibodies as well as peptide-conjugated phosphorodiamidate morpholino oligonucleotides (PPMOs) suppressing the expression of individual PCs are described in detail in Chap. 11. Combinatorial treatment with protease inhibitors and virus-targeting agents such as ribavirin and favipiravir allows a drastic reduction of the drug dosages while maintaining full inhibitory efficacy. Moreover, the application of PC inhibitors prevents the development of resistance to virus-targeting drugs (Lu et al. 2015; Garten et al. 2015).
References
Abifadel M, Varret M, Rabès JP, Allard D, Ouguerram K, Devillers M, Cruaud C, Benjannet S, Wickham L, Erlich D, Derré A, Villéger L, Farnier M, Beucler I, Bruckert E, Chambaz J, Chanu B, Lecerf JM, Luc G, Moulin P, Weissenbach J, Prat A, Krempf M, Junien C, Seidah NG, Boileau C. Mutations in PCSK9 cause autosomal dominant hypercholesterolemia. Nat Genet. 2003;34(2):154–6.
Seidah NG, Abifadel M, Prost S, Boileau C, Prat A. The proprotein convertases in hypercholesterolemia and cardiovascular diseases: emphasis on Proprotein Convertase Subtilisin/Kexin 9. Pharmacol Rev. 2017;69(1):33–52. Review.
Achstetter T, Wolf DH. Hormone processing and membrane-bound proteinases in yeast. EMBO J. 1985;4(1):173–1.
Allison LMC, Salter MWAP, Kiguwa S, Howard CR. Analysis of the glycoprotein gene of Tacaribe virus and neutralization-resistant variants. J Gen Virol. 1991;72:2025–9.
Altamura LA, Bertolotti-Ciarlet A, Teigler J, Paragas J, Schmaljohn CS, Doms RW. Identification of a novel C-terminal cleavage of Crimean-Congo hemorrhagic fever virus PreGN that leads to generation of an NSM protein. J Virol. 2007;81(12):6632–42.
Anders A, Gilbert S, Garten W, Postina R, Fahrenholz F. Regulation of the alpha-secretase ADAM10 by its prodomain and proprotein convertases. FASEB J. 2001;15(10):1837–9.
Anderson ED, Thomas L, Hayflick JS, Thomas G. Inhibition of HIV-1 gp160-dependent membrane fusion by a furin-directed alpha 1-antitrypsin variant. J Biol Chem. 1993;268(33):24887–91.
Anderson ED, VanSlyke JK, Thulin CD, Jean F, Thomas G. Activation of the furin endoprotease is a multiple-step process: requirements for acidification and internal propeptide cleavage. EMBO J. 1997;16(7):1508–18.
Artenstein AW, Opal SM. Proprotein convertases in health and disease. N Engl J Med. 2011;365(26):2507–18. Review.
Assadi M, Sharpe JC, Snell C, Loh YP. The C-terminus of prohormone convertase 2 is sufficient and necessary for Raft association and sorting to the regulated secretory pathway. Biochemistry. 2004;43(24):7798–807.
Barr PJ. Mammalian subtilisins: the long-sought dibasic processing endoproteases. Cell. 1991;66(1):1–3. Review.
Basak A, Touré BB, Lazure C, Mbikay M, Chrétien M, Seidah NG. Enzymic characterization in vitro of recombinant proprotein convertase PC4. Biochem J. 1999;343(Pt 1):29–37.
Basak A, Mitra A, Basak S, Pasko C, Chrétien M, Seaton P. A fluorogenic peptide containing the processing site of human SARS corona virus S-protein: kinetic evaluation and NMR structure elucidation. ChemBioChem. 2007;8(9):1029–37.
Belouzard S, Chu VC, Whittaker GR. Activation of the SARS coronavirus spike protein via sequential proteolytic cleavage at two distinct sites. Proc Natl Acad Sci U S A. 2009;106(14):5871–6. https://doi.org/10.1073/pnas.0809524106. Epub 2009 Mar 24.
Bergeron É, Zivcec M, Chakrabarti AK, Nichol ST, Albariño CG, Spiropoulou CF. Recovery of recombinant Crimean Congo hemorrhagic fever virus reveals a function for non-structural glycoproteins cleavage by Furin. PLoS Pathog. 2015;11(5):e1004879.
Beyer WR, Pöpplau D, Garten W, von Laer D, Lenz O. Endoproteolytic processing of the lymphocytic choriomeningitis virus glycoprotein by the subtilase SKI-1/S1P. Virology. 2003;77(5):2866–72.
Bhattacharjya S, Xu P, Zhong M, Chrétien M, Seidah NG, Ni F. Inhibitory activity and structural characterization of a C-terminal peptide fragment derived from the prosegment of the proprotein convertase PC7. Biochemistry. 2000;39(11):2868–77.
Bodvard K, Mohlin J, Knecht W. Recombinant expression, purification, and kinetic and inhibitor characterisation of human site-1-protease. Protein Expr Purif. 2007;51(2):308–19.
Bosch FX, Garten W, Klenk HD, Rott R. Proteolytic cleavage of influenza virus hemagglutinins: primary structure of the connecting peptide between HA1 and HA2 determines proteolytic cleavability and pathogenicity of Avian influenza viruses. Virology. 1981;113(2):725–35.
Bosch BJ, Bartelink W, Rottier PJ. Cathepsin L functionally cleaves the severe acute respiratory syndrome coronavirus class I fusion protein upstream of rather than adjacent to the fusion peptide. J Virol. 2008;82(17):8887–90.
Bosshart H, Humphrey J, Deignan E, Davidson J, Drazba J, Yuan L, Oorschot V, Peters PJ, Bonifacino JS. The cytoplasmic domain mediates localization of furin to the trans-Golgi network en route to the endosomal/lysosomal system. J Cell Biol. 1994;126(5):1157–72.
Böttcher-Friebertshäuser E, Garten W, Matrosovich M, Klenk HD. The hemagglutinin: a determinant of pathogenicity. Curr Top Microbiol Immunol. 2014;385:3–34. Review.
Braks JA, Martens GJ. 7B2 is a neuroendocrine chaperone that transiently interacts with prohormone convertase PC2 in the secretory pathway. Cell. 1994;78(2):263–73.
Bresnahan PA, Leduc R, Thomas L, Thorner J, Gibson HL, Brake AJ, Barr PJ, Thomas G. Human fur gene encodes a yeast KEX2-like endoprotease that cleaves pro-beta-NGF in vivo. J Cell Biol. 1990;111(6 Pt 2):2851–9.
Brown MS, Goldstein JL. A proteolytic pathway that controls the cholesterol content of membranes, cells, and blood. Proc Natl Acad Sci U S A. 1999;96(20):11041–8. Review.
Brücher KH, Garten W, Klenk HD, Shaw E, Radsak K. Inhibition of endoproteolytic cleavage of cytomegalovirus (HCMV) glycoprotein B by palmitoyl-peptidyl-chloromethyl ketone. Virology. 1990;178(2):617–20.
Bruzzaniti A, Goodge K, Jay P, Taviaux SA, Lam MH, Berta P, Martin TJ, Moseley JM, Gillespie MT. PC8 [corrected], a new member of the convertase family. Biochem J. 1996;314(Pt 3):727–31.
Burri DJ, Pasqual G, Rochat C, Seidah NG, Pasquato A, Kunz S. Molecular characterization of the processing of arenavirus envelope glycoprotein precursors by subtilisin kexin isozyme-1/site-1 protease. J Virol. 2012;86(9):4935–46.
Burri DJ, da Palma JR, Kunz S, Pasquato A. Envelope glycoprotein of arenaviruses. Virus. 2012;4(10):2162–81.
Burri DJ, da Palma JR, Seidah NG, Zanotti G, Cendron L, Pasquato A, Kunz S. Differential recognition of Old World and New World arenavirus envelope glycoproteins by subtilisin kexin isozyme 1 (SKI-1)/site 1 protease (S1P). J Virol. 2013;87(11):6406–14.
Cameron A, Appel J, Houghten RA, Lindberg I. Polyarginines are potent furin inhibitors. Biol Chem. 2000;275(47):36741–9.
Canaff L, Bennett HP, Hou Y, Seidah NG, Hendy GN. Proparathyroid hormone processing by the proprotein convertase-7: comparison with furin and assessment of modulation of parathyroid convertase messenger ribonucleic acid levels by calcium and 1,25-dihydroxyvitamin D3. Endocrinology. 1999;140(8):3633–42.
Cavanagh D, Davis PJ, Pappin DJ, Binns MM, Boursnell ME, Brown TD. Coronavirus IBV: partial amino terminal sequencing of spike polypeptide S2 identifies the sequence Arg-Arg-Phe-Arg-Arg at the cleavage site of the spike precursor propolypeptide of IBV strains Beaudette and M41. Virus Res. 1986;4(2):133–43.
Chandran K, Sullivan NJ, Felbor U, Whelan SP, Cunningham JM. Endosomal proteolysis of the Ebola virus glycoprotein is necessary for infection. Science. 2005;308(5728):1643–5.
Cieplik M, Klenk HD, Garten W. Identification and characterization of spodoptera frugiperda furin: a thermostable subtilisin-like endopeptidase. Biol Chem. 1998;379(12):1433–40.
Constam DB, Calfon M, Robertson EJ. SPC4, SPC6, and the novel protease SPC7 are coexpressed with bone morphogenetic proteins at distinct sites during embryogenesis. J Cell Biol. 1996;134(1):181–91.
Creemers JW, Khatib AM. Knock-out mouse models of proprotein convertases: unique functions or redundancy? Front Biosci. 2008;13:4960–71. Review.
Creemers JW, Vey M, Schäfer W, Ayoubi TA, Roebroek AJ, Klenk HD, Garten W, Van de Ven WJ. Endoproteolytic cleavage of its propeptide is a prerequisite for efficient transport of furin out of the endoplasmic reticulum. J Biol Chem. 1995;270(6):2695–70.
Creemers JWM, Usac EF, Bright NA, Van de Loo JW, Jansen E, Van de Ven WJM, Hutton JC. Identification of a transferable sorting domain for the regulated pathway in the prohormone convertase PC2. J Biol Chem. 1996;271:25284–91.
Creemers JW, van de Loo JW, Plets E, Hendershot LM, Van De Ven WJ. Binding of BiP to the processing enzyme lymphoma proprotein convertase prevents aggregation, but slows down maturation. J Biol Chem. 2000;275(49):38842–7.
Creemers JW, Pritchard LE, Gyte A, Le Rouzic P, Meulemans S, Wardlaw SL, Zhu X, Steiner DF, Davies N, Armstrong D, Lawrence CB, Luckman SM, Schmitz CA, Davies RA, Brennand JC, White A. Agouti-related protein is posttranslationally cleaved by proprotein convertase 1 to generate agouti-related protein (AGRP)83-132: interaction between AGRP83-132 and melanocortin receptors cannot be influenced by syndecan-3. Endocrinology. 2006;147(4):1621–31.
Cunningham D, Danley DE, Geoghegan KF, Griffor MC, Hawkins JL, Subashi TA, Varghese AH, Ammirati MJ, Culp JS, Hoth LR, et al. Structural and biophysical studies of PCSK9 and its mutants linked to familial hypercholesterolemia. Nat Struct Mol Biol. 2007;14:413–9.
da Palma JR, Burri DJ, Oppliger J, Salamina M, Cendron L, de Laureto PP, Seidah NG, Kunz S, Pasquato A. Zymogen activation and subcellular activity of subtilisin kexin isozyme 1/site 1 protease. J Biol Chem. 2014;289:35743–56.
da Palma JR, Cendron L, Seidah NG, Pasquato A, Kunz S. Mechanism of folding and activation of Subtilisin Kexin Isozyme-1 (SKI-1)/Site-1 Protease (S1P). J Biol Chem. 2016;291(5):2055–66.
Dahms SO, Hardes K, Becker GL, Steinmetzer T, Brandstetter H, Than ME. X-ray structures of human furin in complex with competitive inhibitors. ACS Chem Biol. 2014;9(5):1113–8.
Dahms SO, Creemers JW, Schaub Y, Bourenkov GP, Zögg T, Brandstetter H, Than ME. The structure of a furin-antibody complex explains non-competitive inhibition by steric exclusion of substrate conformers. Sci Rep. 2016;6:34303.
Dahms SO, Arciniega M, Steinmetzer T, Huber R, Than ME. Structure of the unliganded form of the proprotein convertase furin suggests activation by a substrate-induced mechanism. Proc Natl Acad Sci U S A. 2016;113(40):11196–201.
Dahms SO, Jiao GS, Than ME. Structural studies revealed active site distortions of human furin by a small molecule inhibitor. ACS Chem Biol. 2017.
Day PM, Lowy DR, Schiller JT. Heparan sulfate-independent cell binding and infection with furin-precleaved papillomavirus capsids. J Virol. 2008;82(24):12565–8.
De Bie I, Savaria D, Roebroek AJ, Day R, Lazure C, Van de Ven WJ, Seidah NG. Processing specificity and biosynthesis of the Drosophila melanogaster convertases dfurin1, dfurin1-CRR, dfurin1-X, and dfurin2. J Biol Chem. 1995;270(3):1020–8.
De Bie I, Marcinkiewicz M, Malide D, Lazure C, Nakyama K, Bendayan M, Seidah NG. The isoforms of proprotein convertase PC5 are sorted to different subcellular compartments. sorting signals YxxL, LL und acidic serine-casein kinase phosphorylation, PC5, PC7, kexin, furin. J Cell Biol. 1996;135:1261–75.
de Haan CA, Stadler K, Godeke GJ, Bosch BJ, Rottier PJ. Cleavage inhibition of the murine coronavirus spike protein by a furin-like enzyme affects cell-cell but not virus-cell fusion. J Virol. 2004;78(11):6048–54.
Declercq J, Meulemans S, Plets E, Creemers JW. I nternalization of proprotein convertase PC7 from plasma membrane is mediated by a novel motif. J Biol Chem. 2012;287(12):9052–60.
Declercq J, Ramos-Molina B, Sannerud R, Brouwers B, Pruniau VPEG, Meulemans S, Plets E, Annaert W, Creemers JWM. Endosome to trans-Golgi network transport of Proprotein Convertase 7 is mediated by a cluster of basic amino acids and palmitoylated cysteines. Eur J Cell Biol. 2017. pii: S0171-9335(16)30198-4.
Decroly E, Benjannet S, Savaria D, Seidah NG. Comparative functional role of PC7 and furin in the processing of the HIV envelope glycoprotein gp160. FEBS Lett. 1997;405(1):68–72.
Dikeakos JD, Di Lello P, Lacombe MJ, Ghirlando R, Legault P, Reudelhuber TL, Omichinski JG. Functional and structural characterization of a dense core secretory granule sorting domain from the PC1/3 protease. Proc Natl Acad Sci U S A. 2009;106(18):7408–13.
Dillon SL, Williamson DM, Elferich J, Radler D, Joshi R, Thomas G, Shinde U. Propeptides are sufficient to regulate organelle-specific pH-dependent activation of furin and proprotein convertase 1/3. J Mol Biol. 2012;423(1):47–62.
Duda A, Stange A, Lüftenegger D, Stanke N, Westphal D, Pietschmann T, Eastman SW, Linial ML, Rethwilm A, Lindemann D. Prototype foamy virus envelope glycoprotein leader peptide processing is mediated by a furin-like cellular protease, but cleavage is not essential for viral infectivity. J Virol. 2004;78(24):13865–70.
Duguay SJ, Lai-Zhang J, Steiner DF. Mutational analysis of the insulin-like growth factor I prohormone processing site. J Biol Chem. 1995;270(29):17566–74.
Eichler R, Lenz O, Garten W, Strecker T. The role of single N-glycans in proteolytic processing and cell surface transport of the Lassa virus glycoprotein GP-C. Virol J. 2006;3:41.
Eickmann M, Kiermayer S, Kraus I, Gössl M, Richt JA, Garten W. Maturation of Borna disease virus glycoprotein. FEBS Lett. 2005;579(21):4751–6.
Eisenberg RJ, Atanasiu D, Cairns TM, Gallagher JR, Krummenacher C, Cohen GH. Herpes virus fusion and entry: a story with many characters. Virus. 2012;4(5):800–32. Review.
Elagoz A, Benjannet S, Mammarbassi A, Wickham L, Seidah NG. Biosynthesis and cellular trafficking of the convertase SKI-1/S1P: ectodomain shedding requires SKI-1 activity. J Biol Chem. 2002;277(13):11265–75.
Elshuber S, Allison SL, Heinz FX, Mandl CW. Cleavage of protein prM is necessary for infection of BHK-21 cells by tick-borne encephalitis virus. J Gen Virol. 2003;84(Pt 1):183–91.
Essalmani R, Zaid A, Marcinkiewicz J, Chamberland A, Pasquato A, Seidah NG, Prat A. In vivo functions of the proprotein convertase PC5/6 during mouse development: Gdf11 is a likely substrate. Proc Natl Acad Sci U S A. 2008;105(15):5750–5.
Essalmani R, Susan-Resiga D, Guillemot J, Kim W, Sachan V, Awan Z, Chamberland A, Asselin MC, Ly K, Desjardins R, Day R, Prat A, Seidah NG. Thrombin activation of protein C requires prior processing by a liver proprotein convertase. J Biol Chem. 2017;292(25):10564–73.
Farooqi IS, Volders K, Stanhope R, Heuschkel R, White A, Lank E, Keogh J, O’Rahilly S, Creemers JW. Hyperphagia and early-onset obesity due to a novel homozygous missense mutation in prohormone convertase 1/3. J Clin Endocrinol Metab. 2007;92(9):3369–73.
Feldmann A, Schäfer MK, Garten W, Klenk HD. Targeted infection of endothelial cells by avian influenza virus A/FPV/Rostock/34 (H7N1) in chicken embryos. J Virol. 2000;74(17):8018–27.
Follis KE, York J, Nunberg JH. Furin cleavage of the SARS coronavirus spike glycoprotein enhances cell-cell fusion but does not affect virion entry. Virology. 2006;350(2):358–69.
Fujii Y, Sakaguchi T, Kiyotani K, Yoshida T. Comparison of substrate specificities against the fusion glycoprotein of virulent Newcastle disease virus between a chick embryo fibroblast processing protease and mammalian subtilisin-like proteases. Microbiol Immunol. 1999;43(2):133–40.
Fuller RS, Brake A, Thorner J. Yeast prohormone processing enzyme (KEX2 gene product) is a Ca2+-dependent serine protease. Proc Natl Acad Sci U S A. 1989a;86(5):1434–8.
Fuller RS, Brake AJ, Thorner J. Intracellular targeting and structural conservation of a prohormone-processing endoprotease. Science. 1989b;246(4929):482–6.
Gabriel G, Garn H, Wegmann M, Renz H, Herwig A, Klenk HD, Stech J. The potential of a protease activation mutant of a highly pathogenic avian influenza virus for a pandemic live vaccine. Vaccine. 2008;26(7):956–65.
Garred O, van Deurs B, Sandvig K. Furin-induced cleavage and activation of Shiga toxin. J Biol Chem. 1995;270(18):10817–21.
Garten W, Bosch FX, Linder D, Rott R, Klenk HD. Proteolytic activation of the influenza virus hemagglutinin: The structure of the cleavage site and the enzymes involved in cleavage. Virology. 1981;115(2):361–74.
Garten W, Linder D, Rott R, Klenk HD. The cleavage site of the hemagglutinin of fowl plague virus. Virology. 1982;122(1):186–90.
Garten W, Kuroda K, Schuy W, Naruse H, Scholtissek C, Klenk HD. Haemagglutinin transport mutants. Vaccine. 1985;3(3 Suppl):227–9.
Garten W, Stieneke A, Shaw E, Wikstrom P, Klenk HD. Inhibition of proteolytic activation of influenza virus hemagglutinin by specific peptidyl chloroalkyl ketones. Virology. 1989;172(1):25–31.
Garten W, Vey M, Ohuchi R, Ohuchi M, Klenk HD. Modification of the cleavage activation of the influenza virus hemagglutinin by site-specific mutagenesis. Behring Inst Mitt. 1991;89:12–22.
Garten W, Hallenberger S, Ortmann D, Schäfer W, Vey M, Angliker H, Shaw E, Klenk HD. Processing of viral glycoproteins by the subtilisin-like endoprotease furin and its inhibition by specific peptidyl chloroalkyl ketones. Biochimie. 1994;76(3-4):217–25. Review
Garten W, Braden C, Arendt A, Peitsch C, Baron J, Lu Y, Pawletko K, Hardes K, Steinmetzer T, Böttcher-Friebertshäuser E. Influenza virus activating host proteases: identification, localization and inhibitors as potential therapeutics. Eur J Cell Biol. 2015;94(7-9):375–83.
Geiselhart V, Bastone P, Kempf T, Schnölzer M, Löchelt M. Furin-mediated cleavage of the feline foamy virus. J Virol. 2004;78(24):13573–81.
Gil-Torregrosa BC, Castano AR, Lopez D, Del Val M. Generation of MHC class I peptide antigens by protein processing in the secretory route by furin. Traffic. 2000;1:641–51.
González-Reyes L, Ruiz-Argüello MB, García-Barreno B, Calder L, López JA, Albar JP, Skehel JJ, Wiley DC, Melero JA. Cleavage of the human respiratory syncytial virus fusion protein at two distinct sites is required for activation of membrane fusion. Proc Natl Acad Sci U S A. 2001;98(17):9859–64.
Gordon VM, Leppla SH. Proteolytic activation of bacterial toxins: role of bacterial and host cell proteases. Infect Immun. 1994;62(2):333–40. Review.
Gordon VM, Rehemtulla A, Leppla SH. A role for PACE4 in the proteolytic activation of anthrax toxin protective antigen. Infect Immun. 1997;65(8):3370–5.
Gorski JP, Huffman NT, Chittur S, Midura RJ, Black C, Oxford J, Seidah NG. Inhibition of proprotein convertase SKI-1 blocks transcription of key extracellular matrix genes regulating osteoblastic mineralization. J Biol Chem. 2011;286(3):1836–49.
Gotoh B, Ohnishi Y, Inocencio NM, Esaki E, Nakayama K, Barr PJ, Thomas G, Nagai Y. Mammalian subtilisin-related proteinases in cleavage activation of the paramyxovirus fusion glycoprotein: superiority of furin/PACE to PC2 or PC1/PC3. J Virol. 1992;66(11):6391–7.
Gu M, Rappaport J, Leppla SH. Furin is important but not essential for the proteolytic maturation of gp160 of HIV-1. FEBS Lett. 1995;365(1):95–7.
Guillemot J, Canuel M, Essalmani R, Prat A, Seidah NG. Implication of the proprotein convertases in iron homeostasis: proprotein convertase 7 sheds human transferrin receptor 1 and furin activates hepcidin. Hepatology. 2013;57(6):2514–24.
Hallenberger S, Bosch V, Angliker H, Shaw E, Klenk HD, Garten W. Inhibition of furin-mediated cleavage activation of HIV-1 glycoprotein gp160. Nature. 1992;360(6402):358–61.
Hallenberger S, Moulard M, Sordel M, Klenk HD, Garten W. The role of eukaryotic subtilisin-like endoproteases for the activation of human immunodeficiency virus glycoproteins in natural host cells. J Virol. 1997;71(2):1036–45.
Hardes K, Becker GL, Lu Y, Dahms SO, Köhler S, Beyer W, Sandvig K, Yamamoto H, Lindberg I, Walz L, von Messling V, Than ME, Garten W, Steinmetzer T. Novel Furin inhibitors with potent anti-infectious activity. ChemMedChem. 2015;10(7):1218–31.
Hardes K, Ivanova T, Thaa B, McInermey GM, Klokk TI, Sandvig K, Kunzel S, Lindberg I, Steinmetzer T. Elongated and shortened peptidomimetic inhibitors of the proprotein convertase furin. ChemMedChem. 2017;12:613–20.
Harrison SC. Viral membrane fusion. Nat Struct Mol Biol. 2008;15(7):690–8.
Hatsuzawa K, Hosaka M, Nakagawa T, Nagase M, Shoda A, Murakami K, Nakayama K. Structure and expression of mouse furin, a yeast Kex2-related protease. Lack of processing of coexpressed prorenin in GH4C1 cells. J Biol Chem. 1990;265(36):22075–8.
Heidner HW, Johnston RE. The amino-terminal residue of Sindbis virus glycoprotein E2 influences virus maturation, specific infectivity for BHK cells, and virulence in mice. J Virol. 1994;68(12):8064–70.
Hendy GN, Bennett HP, Gibbs BF, Lazure C, Day R, Seidah NG. Proparathyroid hormone is preferentially cleaved to parathyroid hormone by the prohormone convertase furin. A mass spectrometric study. J Biol Chem. 1995;270(16):9517–25.
Henrich S, Cameron A, Bourenkov GP, Kiefersauer R, Huber R, Lindberg I, Bode W, Than ME. The crystal structure of the proprotein processing proteinase furin explains its stringent specificity. Nat Struct Biol. 2003;10(7):520–6. PDB: 1PJ8. Erratum in: Nat Struct Biol. 2003;10(8):669.
Henrich S, Lindberg I, Bode W, Than ME. Proprotein convertase models based on the crystal structures of furin and kexin: explanation of their specificity. J Mol Biol. 2005;345(2):211–27.
Himmelspach M, Pfleiderer M, Fischer BE, Plaimauer B, Antoine G, Falkner FG, Dorner F, Schlokat U. Recombinant human factor X: high yield expression and the role of furin in proteolytic maturation in vivo and in vitro. Thromb Res. 2000;97(2):51–67.
Horimoto T, Kawaoka Y. The hemagglutinin cleavability of a virulent avian influenza virus by subtilisin-like endoproteases is influenced by the amino acid immediately downstream of the cleavage site. Virology. 1995;210(2):466–70.
Horimoto T, Nakayama K, Smeekens SP, Kawaoka Y. Proprotein-processing endoproteases PC6 and furin both activate hemagglutinin of virulent avian influenza viruses. J Virol. 1994;68(9):6074–8.
Ito K, Kim KH, Lok AS, Tong S. Characterization of genotype-specific carboxyl-terminal cleavage sites of hepatitis B virus e antigen precursor and identification of furin as the candidate enzyme. J Virol. 2009;83(8):3507–17.
Jain SK, De Candido S, Kielian M. Processing of the p62 envelope precursor protein of Semliki Forest virus. J Biol Chem. 1991;266(9):5756–61.
Jardetzky TS, Lamb RA. Activation of paramyxovirus membrane fusion and virus entry. Curr Opin Virol. 2014;5:24–33. Review.
Jean F, Stella K, Thomas L, Liu G, Xiang Y, Reason AJ, Thomas G. alpha1-Antitrypsin Portland, a bioengineered serpin highly selective for furin: application as an antipathogenic agent. Proc Natl Acad Sci U S A. 1998;95(13):7293–8.
Johannsen E, Luftig M, Chase MR, Weicksel S, Cahir-McFarland E, Illanes D, Sarracino D, Kieff E. Proteins of purified Epstein-Barr virus. Proc Natl Acad Sci U S A. 2004;101(46):16286–91.
Julius D, Brake A, Blair L, Kunisawa R, Thorner J. Isolation of the putative structural gene for the lysine-arginine cleaving endopeptidase required for processing of yeast prepro-alpha-factor. Cell. 1984;37:1075–89.
Kacprzak MM, Peinado JR, Than ME, Appel J, Henrich S, Lipkind G, Houghten RA, Bode W, Lindberg I. Inhibition of furin by polyarginine-containing peptides: nanomolar inhibition by nona-D-arginine. J Biol Chem. 2004;279(35):36788–94.
Kawaoka Y, Webster RG. Sequence requirements for cleavage activation of influenza virus hemagglutinin expressed in mammalian cells. Proc Natl Acad Sci U S A. 1988;85(2):324–8.
Kawaoka Y, Nestorowicz A, Alexander DJ, Webster RG. Molecular analyses of the hemagglutinin genes of H5 influenza viruses: origin of a virulent turkey strain. Virology. 1987;158(1):218–27.
Keelapang P, Sriburi R, Supasa S, Panyadee N, Songjaeng A, Jairungsri A, Puttikhunt C, Kasinrerk W, Malasit P, Sittisombut N. Alterations of pr-M cleavage and virus export in pr-M junction chimeric dengue viruses. J Virol. 2004;78(5):2367–81.
Kemmler W, Peterson JD, Steiner DF. Studies on the conversion of proinsulin to insulin. I. Conversion in vitro with trypsin and carboxypeptidase B. J Biol Chem. 1971;246(22):6786–91.
Khatib AM, Siegfried G, Prat A, Luis J, Chrétien M, Metrakos P, Seidah NG. Inhibition of proprotein convertases is associated with loss of growth and tumorigenicity of HT-29 human colon carcinoma cells: importance of insulin-like growth factor-1 (IGF-1) receptor processing in IGF-1-mediated functions. J Biol Chem. 2001;276(33):30686–93.
Kiefer MC, Tucker JE, Joh R, Landsberg KE, Saltman D, Barr PJ. Identification of a second human subtilisin-like protease gene in the fes/fps region of chromosome 15. DNA Cell Biol. 1991;10(10):757–69.
Klenk HD, Garten W. Activation cleavage of viral spike proteins by host proteases. In: Wimmer E, editor. Cellular receptors for animal viruses. Cold Spring Harbor: Cold Spring Harbor Laboratory Press; 1994. p. 241–80.
Klenk HD, Garten W, Rott R. Inhibition of proteolytic cleavage of the hemagglutinin of influenza virus by the calcium-specific ionophore A23187. EMBO J. 1984;3(12):2911–5.
Kopp A, Blewett E, Misra V, Mettenleiter TC. Proteolytic cleavage of bovine herpesvirus 1 (BHV-1) glycoprotein gB is not necessary for its function in BHV-1 or pseudorabies virus. J Virol. 1994;68(3):1667–74.
Kouretova J, Hammamy MZ, Epp A, Hardes K, Kallis S, Zhang L, Hilgenfeld R, Bartenschlager R, Steinmetzer T. Effects of NS2B-NS3 protease and furin inhibition on West Nile and Dengue virus replication. J Enzyme Inhib Med Chem. 2017;32(1):712–21.
Kuno G, Chang GJ. Full-length sequencing and genomic characterization of Bagaza, Kedougou, and Zika viruses. Arch Virol. 2007;152(4):687–96.
Kunz S, Edelmann KH, de la Torre JC, Gorney R, Oldstone MB. Mechanisms for lymphocytic choriomeningitis virus glycoprotein cleavage, transport, and incorporation into virions. Virology. 2003;314(1):168–78.
Kuroda K, Hauser C, Rott R, Klenk HD, Doerfler W. Expression of the influenza virus haemagglutinin in insect cells by a baculovirus vector. EMBO J. 1986;5(6):1359–65.
Kuroda K, Gröner A, Frese K, Drenckhahn D, Hauser C, Rott R, Doerfler W, Klenk HD. Synthesis of biologically active influenza virus hemagglutinin in insect larvae. J Virol. 1989;63(4):1677–85.
Le QT, Blanchet M, Seidah NG, Labonté P. Plasma Membrane Tetraspanin CD81 complexes with Proprotein Convertase Subtilisin/Kexin Type 9 (PCSK9) and low density lipoprotein receptor (LDLR), and its levels are reduced by PCSK9. J Biol Chem. 2015;290(38):23385–400.
Leduc R, Molloy SS, Thorne BA, Thomas G. Activation of human furin precursor processing endoprotease occurs by an intramolecular autoproteolytic cleavage. J Biol Chem. 1992;267(20):14304–8.
Lee SN, Lindberg I. 7B2 prevents unfolding and aggregation of prohormone convertase. Endocrinology. 2008;149(8):4116–27.
Lee SN, Lee DH, Lee MG, Yoon JH. Proprotein convertase 5/6a is associated with bone morphogenetic protein-2-induced squamous cell differentiation. Am J Respir Cell Mol Biol. 2015;52(6):749–61.
Lenz O, ter Meulen J, Feldmann H, Klenk HD, Garten W. Identification of a novel consensus sequence at the cleavage site of the Lassa virus glycoprotein. J Virol. 2000;74(23):11418–21.
Lenz O, ter Meulen J, Klenk HD, Seidah NG, Garten W. The Lassa virus glycoprotein precursor GP-C is proteolytically processed by subtilase SKI-1/S1P. Proc Natl Acad Sci U S A. 2001;98(22):12701–5. Epub 2001 Oct 16.
Li S, Sun Z, Pryce R, Parsy ML, Fehling SK, Schlie K, Siebert CA, Garten W, Bowden TA, Strecker T, Huiskonen JT. Acidic pH-induced conformations and LAMP1 binding of the Lassa Virus glycoprotein spike. PLoS Pathog. 2016;12(2):e1005418.
Long G, Pan X, Westenberg M, Vlak JM. Functional role of the cytoplasmic tail domain of the major envelope fusion protein of group II baculoviruses. J Virol. 2006;80(22):11226–34.
Longpré JM, Leduc R. Identification of prodomain determinants involved in ADAMTS-1 biosynthesis. J Biol Chem. 2004;279(32):33237–45.
Lopez-Perez E, Zhang Y, Frank SJ, Creemers J, Seidah N, Checler F. Constitutive alpha-secretase cleavage of the beta-amyloid precursor protein in the furin-deficient LoVo cell line: involvement of the pro-hormone convertase 7 and the disintegrin metalloprotease ADAM10. J Neurochem. 2001;76(5):1532–9.
Lu Y, Hardes K, Dahms SO, Böttcher-Friebertshäuser E, Steinmetzer T, Than ME, Klenk HD, Garten W. Peptidomimetic furin inhibitor MI-701 in combination with oseltamivir and ribavirin efficiently blocks propagation of highly pathogenic avian influenza viruses and delays high level oseltamivir resistance in MDCK cells. Antiviral Res. 2015;120:89–100.
Lusson J, Vieau D, Hamelin J, Day R, Chrétien M, Seidah NG. cDNA structure of the mouse and rat subtilisin/kexin-like PC5: a candidate proprotein convertase expressed in endocrine and nonendocrine cells. Proc Natl Acad Sci U S A. 1993;90(14):6691–5.
Mains RE, Berard CA, Denault JB, Zhou A, Johnson RC, Leduc R. PACE4: a subtilisin-like endoprotease with unique properties. Biochem J. 1997;321(Pt 3):587–93.
Maisa A, Ströher U, Klenk HD, Garten W, Strecker T. Inhibition of Lassa virus glycoprotein cleavage and multicycle replication by site 1 protease-adapted alpha(1)-antitrypsin variants. PLoS Negl Trop Dis. 2009;3(6):e446.
Mbikay M, Tadros H, Ishida N, Lerner CP, De Lamirande E, Chen A, El-Alfy M, Clermont Y, Seidah NG, Chrétien M, Gagnon C, Simpson EM. Impaired fertility in mice deficient for the testicular germ-cell protease PC4. Proc Natl Acad Sci U S A. 1997;94(13):6842–6.
McCune JM, Rabin LB, Feinberg MB, Lieberman M, Kosek JC, Reyes GR, Weissman IL. Endoproteolytic cleavage of gp160 is required for the activation of human immunodeficiency virus. Cell. 1988;53(1):55–67.
Meerabux J, Yaspo ML, Roebroek AJ, Van de Ven WJ, Lister TA, Young BD. A new member of the proprotein convertase gene family (LPC) is located at a chromosome translocation breakpoint in lymphomas. Cancer Res. 1996;56(3):448–51.
Millet JK, Whittaker GR. Host cell entry of Middle East respiratory syndrome coronavirus after two-step, furin-mediated activation of the spike protein. Proc Natl Acad Sci U S A. 2014;111:15214–9.
Millet JK, Whittaker GR. Host cell proteases: critical determinants of coronavirus tropism and pathogenesis. Virus Res. 2015;202:120–34.
Misumi Y, Ohkubo K, Sohda M, Takami N, Oda K, Ikehara Y. Intracellular processing of complement pro-C3 and proalbumin is inhibited by rat alpha 1-protease inhibitor variant (Met352----Arg) in transfected cells. Biochem Biophys Res Commun. 1990;171(1):236–42.
Misumi Y, Sohda M, Ikehara Y. Sequence of the cDNA encoding rat furin, a possible propeptide-processing endoprotease. Nucleic Acids Res. 1990;18(22):6719.
Mizuno K, Nakamura T, Matsuo H. A unique membrane-bound, calcium-dependent endopeptidase with specificity toward paired basic residues in rat liver Golgi fractions. Biochem Biophys Res Commun. 1989;164(2):780–7.
Moehring JM, Inocencio NM, Robertson BJ, Moehring TJ. Expression of mouse furin in a Chinese hamster cell resistant to Pseudomonas exotoxin A and viruses complements the genetic lesion. J Biol Chem. 1993;268(4):2590–4.
Molloy SS, Bresnahan PA, Leppla SH, Klimpel KR, Thomas G. Human furin is a calcium-dependent serine endoprotease that recognizes the sequence Arg-X-X-Arg and efficiently cleaves anthrax toxin protective antigen. J Biol Chem. 1992;267(23):16396–402.
Molloy SS, Anderson ED, Jean F, Thomas G. Bi-cycling the furin pathway: from TGN localization to pathogen activation and embryogenesis. Trends Cell Biol. 1999;9(1):28–35. Review.
Mori K, Imamaki A, Nagata K, Yonetomi Y, Kiyokage-Yoshimoto R, Martin TJ, Gillespie MT, Nagahama M, Tsuji A, Matsuda Y. Subtilisin-like proprotein convertases, PACE4 and PC8, as well as furin, are endogenous proalbumin convertases in HepG2 cells. J Biochem. 1999;125(3):627–33.
Morikawa Y, Barsov E, Jones I. Legitimate and illegitimate cleavage of human immunodeficiency virus glycoproteins by furin. J Virol. 1993;67(6):3601–4.
Munzer JS, Basak A, Zhong M, Mamarbachi A, Hamelin J, Savaria D, Lazure C, Hendy GN, Benjannet S, Chrétien M, Seidah NG. In vitro characterization of the novel proprotein convertase PC7. J Biol Chem. 1997;272(32):19672–81.
Nagai Y. Virus activation by host proteinases. A pivotal role in the spread of infection, tissue tropism and pathogenicity. Microbiol Immunol. 1995;39(1):1–9. Review.
Nakagawa T, Hosaka M, Torii S, Watanabe T, Murakami K, Nakayama K. Identification and functional expression of a new member of the mammalian Kex2-like processing endoprotease family: its striking structural similarity to PACE4. J Biochem. 1993;113(2):132–5.
Nakagawa T, Suzuki-Nakagawa C, Watanabe A, Asami E, Matsumoto M, Nakano M, Ebihara A, Uddin MN, Suzuki F. Site-1 protease is required for the generation of soluble (pro)renin receptor. J Biochem. 2017;161(4):369–79.
Nakayama K. Furin: a mammalian subtilisin/Kex2p-like endoprotease involved in processing of a wide variety of precursor proteins. Biochem J. 1997;327(Pt 3):625–35.
Nakayama K, Kim WS, Torii S, Hosaka M, Nakagawa T, Ikemizu J, Baba T, Murakami K. Identification of the fourth member of the mammalian endoprotease family homologous to the yeast Kex2 protease. Its testis-specific expression. J Biol Chem. 1992;267(9):5897–900.
Neumann G, Horimoto T, Kawaoka Y. Reverse genetics of influenza viruses–applications in research and vaccine design. In: Klenk H-D, Matrosovich MN, Stech J, editors. Avian Influenza. Monogr Virol, vol. 27. Basel: Karger; 2008. p. 118–33.
Nour N, Basak A, Chrétien M, Seidah NG. Structure-function analysis of the prosegment of the proprotein convertase PC5A. J Biol Chem. 2003;278(5):2886–95.
Nour N, Mayer G, Mort JS, Salvas A, Mbikay M, Morrison CJ, Overall CM, Seidah NG. The cysteine-rich domain of the secreted proprotein convertases PC5A and PACE4 functions as a cell surface anchor and interacts with tissue inhibitors of metalloproteinases. Mol Biol Cell. 2005;16(11):5215–26.
Oda K, Misumi Y, Ikehara Y, Brennan SO, Hatsuzawa K, Nakayama K. Proteolytic cleavages of proalbumin and complement Pro-C3 in vitro by a truncated soluble form of furin, a mammalian homologue of the yeast Kex2 protease. Biochem Biophys Res Commun. 1992;189(3):1353–61.
Ohnishi Y, Shioda T, Nakayama K, Iwata S, Gotoh B, Hamaguchi M, Nagai Y. A furin-defective cell line is able to process correctly the gp160 of human immunodeficiency virus type 1. J Virol. 1994;68(6):4075–9.
Oliver SL, Sommer M, Zerboni L, Rajamani J, Grose C, Arvin AM. Mutagenesis of varicella-zoster virus glycoprotein B: putative fusion loop residues are essential for viral replication, and the furin cleavage motif contributes to pathogenesis in skin tissue in vivo. J Virol. 2009;83(15):7495–506.
Olmstead AD, Knecht W, Lazarov I, Dixit SB, Jean F. Human subtilase SKI-1/S1P is a master regulator of the HCV Lifecycle and a potential host cell target for developing indirect-acting antiviral agents. PLoS Pathog. 2012;8(1):e1002468.
Orlich M, Linder D, Rott R. Trypsin-resistant protease activation mutants of an influenza virus. J Gen Virol. 1995;76(Pt 3):625–33.
Ortmann D, Ohuchi M, Angliker H, Shaw E, Garten W, Klenk HD. Proteolytic cleavage of wild type and mutants of the F protein of human parainfluenza virus type 3 by two subtilisin-like endoproteases, furin and Kex2. J Virol. 1994;68(4):2772–6.
Ozden S, Lucas-Hourani M, Ceccaldi PE, Basak A, Valentine M, Benjannet S, Hamelin J, Jacob Y, Mamchaoui K, Mouly V, Desprès P, Gessain A, Butler-Browne G, Chrétien M, Tangy F, Vidalain PO, Seidah NG. Inhibition of Chikungunya virus infection in cultured human muscle cells by furin inhibitors: impairment of the maturation of the E2 surface glycoprotein. J Biol Chem. 2008;283(32):21899–908.
Pan H, Che FY, Peng B, Steiner DF, Pintar JE, Fricker LD. The role of prohormone convertase-2 in hypothalamic neuropeptide processing: a quantitative neuropeptidomic study. J Neurochem. 2006;98(6):1763–77.
Pang YJ, Tan XJ, Li DM, Zheng ZH, Lei RX, Peng XM. Therapeutic potential of furin inhibitors for the chronic infection of hepatitis B virus. Liver Int. 2013;33(8):1230–8.
Paquet L, Bergeron F, Boudreault A, Seidah NG, Chrétien M, Mbikay M, Lazure C. The neuroendocrine precursor 7B2 is a sulfated protein proteolytically processed by a ubiquitous furin-like convertase. J Biol Chem. 1994;269(30):19279–85.
Pei D, Weiss SJ. Furin-dependent intracellular activation of the human stromelysin-3 zymogen. Nature. 1995;375(6528):244–7.
Piper DE, Jackson S, Liu Q, Romanow WG, Shetterly S, Thibault ST, Shan B, Walker NP. The crystal structure of PCSK9: a regulator of plasma LDL-cholesterol. Structure. 2007;15(5):545–52.
Plaimauer B, Mohr G, Wernhart W, Himmelspach M, Dorner F, Schlokat U. ‘Shed’ furin: mapping of the cleavage determinants and identification of its C-terminus. Biochem J. 2001;354(Pt 3):689–95.
Porter AG, Barber C, Carey NH, Hallewell RA, Threlfall G, Emtage JS. Complete nucleotide sequence of an influenza virus haemagglutinin gene from cloned DNA. Nature. 1979;282(5738):471–7.
Pritzer E, Kuroda K, Garten W, Nagai Y, Klenk HD. A host range mutant of Newcastle disease virus with an altered cleavage site for proteolytic activation of the F protein. Virus Res. 1990;15(3):237–42.
Puente XS, Sánchez LM, Overall CM, López-Otín C. Human and mouse proteases: a comparative genomic approach. Nat Rev Genet. 2003;4(7):544–58. Review.
Puthavathana P, Auewarakul P, Charoenying PC, Sangsiriwut K, Pooruk P, Boonnak K, Khanyok R, Thawachsupa P, Kijphati R, Sawanpanyalert P. Molecular characterization of the complete genome of human influenza H5N1 virus isolates from Thailand. J Gen Virol. 2005;86(Pt 2):423–33.
Qiu Q, Basak A, Mbikay M, Tsang BK, Gruslin A. Role of pro-IGF-II processing by proprotein convertase 4 in human placental development. Proc Natl Acad Sci U S A. 2005;102(31):11047–52.
Ramos-Molina B, Lindberg I. Phosphorylation and alternative splicing of 7B2 reduce Prohormone Convertase 2 activation. Mol Endocrinol. 2015;29(5):756–64.
Rehemtulla A, Barr PJ, Rhodes CJ, Kaufman RJ. PACE4 is a member of the mammalian propeptidase family that has overlapping but not identical substrate specificity to PACE. Biochemistry. 1993;32(43):11586–90.
Remacle AG, Shiryaev SA, Oh ES, Cieplak P, Srinivasan A, Wei G, Liddington RC, Ratnikov BI, Parent A, Desjardins R, Day R, Smith JW, Lebl M, Strongin AY. Substrate cleavage analysis of furin and related proprotein convertases. A comparative study. J Biol Chem. 2008;283(30):20897–906.
Rice CM, Lenches EM, Eddy SR, Shin SJ, Sheets RL, Strauss JH. Nucleotide sequence of yellow fever virus: implications for flavivirus gene expression and evolution. Science. 1985;229(4715):726–33.
Richards RM, Lowy DR, Schiller JT, Day PM. Cleavage of the papillomavirus minor capsid protein, L2, at a furin consensus site is necessary for infection. Proc Natl Acad Sci U S A. 2006;103:1522–7.
Richardson C, Hull D, Greer P, Hasel K, Berkovich A, Englund G, Bellini W, Rima B, Lazzarini R. The nucleotide sequence of the mRNA encoding the fusion protein of measles virus (Edmonston strain): a comparison of fusion proteins from several different paramyxoviruses. Virology. 1986;155(2):508–23.
Richt JA, Fürbringer T, Koch A, Pfeuffer I, Herden C, Bause-Niedrig I, Garten W. Processing of the Borna disease virus glycoprotein gp94 by the subtilisin-like endoprotease furin. J Virol. 1998;72(5):4528–33.
Robertson BJ, Moehring JM, Moehring TJ. Defective processing of the insulin receptor in an endoprotease-deficient Chinese hamster cell strain is corrected by expression of mouse furin. J Biol Chem. 1993;268(32):24274–7.
Rockwell NC, Krysan DJ, Komiyama T, Fuller RS. Precursor processing by kex2/furin proteases. Chem Rev. 2002;102(12):4525–48.
Roebroek AJ, Schalken JA, Bussemakers MJ, van Heerikhuizen H, Onnekink C, Debruyne FM, Bloemers HP, Van de Ven WJ. Characterization of human c-fes/fps reveals a new transcription unit (fur) in the immediately upstream region of the proto-oncogene. Mol Biol Rep. 1986;11(2):117–25.
Roebroek AJ, Pauli IG, Zhang Y, van de Ven WJ. cDNA sequence of a Drosophila melanogaster gene, Dfur1, encoding a protein structurally related to the subtilisin-like proprotein processing enzyme furin. FEBS Lett. 1991;289(2):133–7.
Roebroek AJ, Umans L, Pauli IG, Robertson EJ, van Leuven F, Van de Ven WJ, Constam DB. Failure of ventral closure and axial rotation in embryos lacking the proprotein convertase Furin. Development. 1998;125(24):4863–76.
Roebroek AJ, Taylor NA, Louagie E, Pauli I, Smeijers L, Snellinx A, Lauwers A, Van de Ven WJ, Hartmann D, Creemers JW. Limited redundancy of the proprotein convertase furin in mouse liver. J Biol Chem. 2004;279(51):53442–50.
Rousselet E, Benjannet S, Hamelin J, Canuel M, Seidah NG. The proprotein convertase PC7: unique zymogen activation and trafficking pathways. J Biol Chem. 2011;286(4):2728–38.
Sakaguchi T, Fujii Y, Kiyotani K, Yoshida T. Correlation of proteolytic cleavage of F protein precursors in paramyxoviruses with expression of the fur, PACE4 and PC6 genes in mammalian cells. J Gen Virol. 1994;75(Pt 10):2821–7.
Sakai J, Rawson RB, Espenshade PJ, Cheng D, Seegmiller AC, Goldstein JL, Brown MS. Molecular identification of the sterol-regulated luminal protease that cleaves SREBPs and controls lipid composition of animal cells. Mol Cell. 1998;2(4):505–14.
Sanchez AJ, Vincent MJ, Nichol ST. Characterization of the glycoproteins of Crimean-Congo hemorrhagic fever virus. J Virol. 2002;76:7263–75.
Sanchez AJ, Vincent MJ, Erickson BR, Nichol ST. Crimean-congo hemorrhagic fever virus glycoprotein precursor is cleaved by Furin-like and SKI-1 proteases to generate a novel 38-kilodalton glycoprotein. J Virol. 2006;80(1):514–25.
Sariola M, Saraste J, Kuismanen E. Communication of post-Golgi elements with early endocytic pathway: regulation of endoproteolytic cleavage of Semliki Forest virus p62 precursor. J Cell Sci. 1995;108(Pt 6):2465–75.
Sato H, Kinoshita T, Takino T, Nakayama K, Seiki M. Activation of a recombinant membrane type 1-matrix metalloproteinase (MT1-MMP) by furin and its interaction with tissue inhibitor of metalloproteinases (TIMP)-2. FEBS Lett. 1996;393(1):101–4.
Scamuffa N, Calvo F, Chrétien M, Seidah NG, Khatib AM. Proprotein convertases: lessons from knockouts. FASEB J. 2006;20(12):1954–63. Review.
Schäfer W, Stroh A, Berghöfer S, Seiler J, Vey M, Kruse ML, Kern HF, Klenk HD, Garten W. Two independent targeting signals in the cytoplasmic domain determine trans-Golgi network localization and endosomal trafficking of the proprotein convertase furin. EMBO J. 1995;14(11):2424–35.
Scheid A, Choppin PW. Protease activation mutants of sendai virus. Activation of biological properties by specific proteases. Virology. 1976;69(1):265–77.
Schlie K, Maisa A, Lennartz F, Ströher U, Garten W, Strecker T. Characterization of Lassa virus glycoprotein oligomerization and influence of cholesterol on virus replication. J Virol. 2010;84(2):983–92.
Schlie K, Strecker T, Garten W. Maturation cleavage within the ectodomain of Lassa virus glycoprotein relies on stabilization by the cytoplasmic tail. FEBS Lett. 2010;584(21):4379–82.
Schlombs K, Wagner T, Scheel J. Site-1 protease is required for cartilage development in zebrafish. Proc Natl Acad Sci U S A. 2003;100(24):14024–9.
Schmidt I, Skinner M, Siddell S. Nucleotide sequence of the gene encoding the surface projection glycoprotein of coronavirus MHV-JHM. J Gen Virol. 1987;68(Pt 1):47–56.
Seidah NG. What lies ahead for the proprotein convertases? Ann N Y Acad Sci. 2011;1220:149–61. Review.
Seidah NG, Prat A. Precursor convertases in the secretory pathway, cytosol and extracellular milieu. Essays Biochem. 2002;38:79–94. Review.
Seidah NG, Prat A. The biology and therapeutic targeting of the proprotein convertases. Nat Rev Drug Discov. 2012;11(5):367–83. Review.
Seidah NG, Gaspar L, Mion P, Marcinkiewicz M, Mbikay M, Chrétien M. cDNA sequence of two distinct pituitary proteins homologous to Kex2 and furin gene products: tissue-specific mRNAs encoding candidates for pro-hormone processing proteinases. DNA Cell Biol. 1990;9(6):415–24. Erratum in: DNA Cell Biol. 1990;9(10):789.
Seidah NG, Day R, Marcinkiewicz M, Benjannet S, Chrétien M. Mammalian neural and endocrine pro-protein and pro-hormone convertases belonging to the subtilisin family of serine proteinases. Enzyme. 1991;45(5-6):271–84. Review.
Seidah NG, Day R, Hamelin J, Gaspar A, Collard MW, Chrétien M. Testicular expression of PC4 in the rat: molecular diversity of a novel germ cell-specific Kex2/subtilisin-like proprotein convertase. Mol Endocrinol. 1992;6(10):1559–70.
Seidah NG, Benjannet S, Pareek S, Chrétien M, Murphy RA. Cellular processing of the neurotrophin precursors of NT3 and BDNF by the mammalian proprotein convertases. FEBS Lett. 1996;379(3):247–50.
Seidah NG, Mowla SJ, Hamelin J, Mamarbachi AM, Benjannet S, Touré BB, Basak A, Munzer JS, Marcinkiewicz J, Zhong M, Barale JC, Lazure C, Murphy RA, Chrétien M, Marcinkiewicz M. Mammalian subtilisin/kexin isozyme SKI-1: a widely expressed proprotein convertase with a unique cleavage specificity and cellular localization. Proc Natl Acad Sci U S A. 1999;96(4):1321–6.
Seidah NG, Benjannet S, Wickham L, Marcinkiewicz J, Jasmin SB, Stifani S, Basak A, Prat A, Chretien M. The secretory proprotein convertase neural apoptosis-regulated convertase 1 (NARC-1): liver regeneration and neuronal differentiation. Proc Natl Acad Sci U S A. 2003;100(3):928–33.
Seidah NG, Mayer G, Zaid A, Rousselet E, Nassoury N, Poirier S, Essalmani R, Prat A. The activation and physiological functions of the proprotein convertases. Int J Biochem Cell Biol. 2008;40(6-7):1111–25. Review.
Seidah NG, Sadr MS, Chrétien M, Mbikay M. The multifaceted proprotein convertases: their unique, redundant, complementary, and opposite functions. J Biol Chem. 2013;288(30):21473–81.
Seidah NG, Awan Z, Chrétien M, Mbikay M. PCSK9: a key modulator of cardiovascular health. Circ Res. 2014;114(6):1022–36. Review.
Shinde U, Thomas G. Insights from bacterial subtilases into the mechanisms of intramolecular chaperone-mediated activation of furin. Methods Mol Biol. 2011;768:59–106. Review.
Shiryaev SA, Chernov AV, Golubkov VS, Thomsen E, Chudin E, Chee M, Kozlov I, Strongin AY, Cieplak P. High-resolution analysis and functional mapping of cleavage sites and substrate proteins of furin in the human proteome. PLoS One. 2013;8:e54290.
Siezen RJ, Creemers JW, Van de Ven WJ. Homology modelling of the catalytic domain of human furin. A model for the eukaryotic subtilisin-like proprotein convertases. Eur J Biochem. 1994;222(2):255–66.
Smeekens SP, Steiner DF. Identification of a human insulinoma cDNA encoding a novel mammalian protein structurally related to the yeast dibasic processing protease Kex2. J Biol Chem. 1990;265(6):2997–3000.
Smeekens SP, Avruch AS, LaMendola J, Chan SJ, Steiner DF. Identification of a cDNA encoding a second putative prohormone convertase related to PC2 in AtT20 cells and islets of Langerhans. Proc Natl Acad Sci U S A. 1991;88(2):340–4.
Sorem J, Longnecker R. Cleavage of Epstein-Barr virus glycoprotein B is required for full function in cell-cell fusion with both epithelial and B cells. J Gen Virol. 2009;90(Pt 3):591–5.
Spiesschaert B, Stephanowitz H, Krause E, Osterrieder N, Azab W. Glycoprotein B of equine herpesvirus type 1 has two recognition sites for subtilisin-like proteases that are cleaved by furin. J Gen Virol. 2016;97(5):1218–28.
Stadler K, Allison SL, Schalich J, Heinz FX. Proteolytic activation of tick-borne encephalitis virus by furin. J Virol. 1997;71:8475–81.
Stawowy P, Kallisch H, Borges Pereira Stawowy N, Stibenz D, Veinot JP, Gräfe M, Seidah NG, Chrétien M, Fleck E, Graf K. Immunohistochemical localization of subtilisin/kexin-like proprotein convertases in human atherosclerosis. Virchows Arch. 2005;446(4):351–9.
Stech J, Garn H, Wegmann M, Wagner R, Klenk HD. A new approach to an influenza live vaccine: modification of the cleavage site of hemagglutinin. Nat Med. 2005;11(6):683–9.
Steiner DF. The proprotein convertases. Curr Opin Chem Biol. 1998;2:31–9.
Steiner DF, Cunningham D, Spigelman L, Aten B. Insulin biosynthesis: evidence for a precursor. Science. 1967;157(3789):697–700.
Stieneke-Gröber A, Vey M, Angliker H, Shaw E, Thomas G, Roberts C, Klenk HD, Garten W. Influenza virus hemagglutinin with multibasic cleavage site is activated by furin, a subtilisin-like endoprotease. EMBO J. 1992;11(7):2407–14.
Strauss EG, Rice CM, Strauss JH. Complete nucleotide sequence of the genomic RNA of Sindbis virus. Virology. 1984;133(1):92–110.
Stroh A, Schäfer W, Berghöfer S, Eickmann M, Teuchert M, Bürger I, Klenk HD, Garten W. A mono phenylalanine-based motif (F790) and a leucine-dependent motif (LI760) mediate internalization of furin. Eur J Cell Biol. 1999;78(3):151–60.
Sturman LS, Holmes KV. Proteolytic cleavage of peplomeric glycoprotein E2 of MHV yields two 90K subunits and activates cell fusion. Adv Exp Med Biol. 1984;173:25–35.
Sucic JF, Moehring JM, Inocencio NM, Luchini JW, Moehring TJ. Endoprotease PACE4 is Ca2+-dependent and temperature-sensitive and can partly rescue the phenotype of a furin-deficient cell strain. Biochem J. 1999;339(Pt 3):639–47.
Takahashi S, Nakagawa T, Banno T, Watanabe T, Murakami K, Nakayama K. Localization of furin to the trans-Golgi network and recycling from the cell surface involves Ser and Tyr residues within the cytoplasmic domain. J Biol Chem. 1995;270(47):28397–401.
Tangrea MA, Bryan PN, Sari N, Orban J. Solution structure of the pro-hormone convertase 1 pro-domain from Mus musculus. J Mol Biol. 2002;320(4):801–12.
Taylor NA, Van De Ven WJ, Creemers JW. Curbing activation: proprotein convertases in homeostasis and pathology. FASEB J. 2003;17(10):1215–27. Review.
Teuchert M, Schäfer W, Berghöfer S, Hoflack B, Klenk HD, Garten W. Sorting of furin at the trans-Golgi network. Interaction of the cytoplasmic tail sorting signals with AP-1 Golgi-specific assembly proteins. J Biol Chem. 1999;274(12):8199–207.
Teuchert M, Berghöfer S, Klenk HD, Garten W. Recycling of furin from the plasma membrane. Functional importance of the cytoplasmic tail sorting signals and interaction with the AP-2 adaptor medium chain subunit. J Biol Chem. 1999;274(51):36781–9.
Than ME, Henrich S, Bourenkov GP, Bartunik HD, Huber R, Bode W. The endoproteinase furin contains two essential Ca2+ ions stabilizing its N-terminus and the unique S1 specificity pocket. Acta Crystallogr D Biol Crystallogr. 2005;61(Pt 5):505–12.
Thomas G. Furin at the cutting edge: from protein traffic to embryogenesis and disease. Nat Rev Mol Cell Biol. 2002;3(10):753–66. Review.
Thomas G, Thorne BA, Thomas L, Allen RG, Hruby DE, Fuller R, Thorner J. Yeast KEX2 endopeptidase correctly cleaves a neuroendocrine prohormone in mammalian cells. Science. 1988;241(4862):226–30.
Toyoda T, Sakaguchi T, Imai K, Inocencio NM, Gotoh B, Hamaguchi M, Nagai Y. Structural comparison of the cleavage-activation site of the fusion glycoprotein between virulent and avirulent strains of Newcastle disease virus. Virology. 1987;158(1):242–7.
Tsuji A, Sakurai K, Kiyokage E, Yamazaki T, Koide S, Toida K, Ishimura K, Matsuda Y. Secretory proprotein convertases PACE4 and PC6A are heparin-binding proteins which are localized in the extracellular matrix. Potential role of PACE4 in the activation of proproteins in the extracellular matrix. Biochim Biophys Acta. 2003;1645(1):95–104.
Tsuneoka M, Nakayama K, Hatsuzawa K, Komada M, Kitamura N, Mekada E. Evidence for involvement of furin in cleavage and activation of diphtheria toxin. J Biol Chem. 1993;268(35):26461–5.
Turpeinen H, Oksanen A, Kivinen V, Kukkurainen S, Uusimäki A, Rämet M, Parikka M, Hytönen VP, Nykter M, Pesu M. Proprotein convertase subtilisin/kexin type 7 (PCSK7) is essential for the zebrafish development and bioavailability of transforming growth factor β1a (TGFβ1a). J Biol Chem. 2013;288(51):36610–23. https://doi.org/10.1074/jbc.M113.453183.
van de Loo JW, Teuchert M, Pauli I, Plets E, Van de Ven WJ, Creemers JW. Dynamic palmitoylation of lymphoma proprotein convertase prolongs its half-life, but is not essential for trans-Golgi network localization. Biochem J. 2000;352(Pt 3):827–33.
van de Ven WJ, Voorberg J, Fontijn R, Pannekoek H, van den Ouweland AM, van Duijnhoven HL, Roebroek AJ, Siezen RJ. Furin is a subtilisin-like proprotein processing enzyme in higher eukaryotes. Mol Biol Rep. 1990;14(4):265–75.
Vey M, Orlich M, Adler S, Klenk HD, Rott R, Garten W. Hemagglutinin activation of pathogenic avian influenza viruses of serotype H7 requires the protease recognition motif R-X-K/R-R. Virology. 1992;188(1):408–13.
Vey M, Schäfer W, Berghöfer S, Klenk HD, Garten W. Maturation of the trans-Golgi network protease furin: compartmentalization of propeptide removal, substrate cleavage, and COOH-terminal truncation. J Cell Biol. 1994;127(6 Pt 2):1829–42.
Vey M, Schäfer W, Reis B, Ohuchi R, Britt W, Garten W, Klenk HD, Radsak K. Proteolytic processing of human cytomegalovirus glycoprotein B (gpUL55) is mediated by the human endoprotease furin. Virology. 1995;206(1):746–9.
Vincent MJ, Sanchez AJ, Erickson BR, Basak A, Chretien M, Seidah NG, Nichol ST. Crimean-Congo hemorrhagic fever virus glycoprotein proteolytic processing by subtilase SKI-1. J Virol. 2003;77(16):8640–9.
Volchkov VE, Feldmann H, Volchkova VA, Klenk HD. Processing of the Ebola virus glycoprotein by the proprotein convertase furin. Proc Natl Acad Sci U S A. 1998;95(10):5762–7.
Volchkov VE, Volchkova VA, Ströher U, Becker S, Dolnik O, Cieplik M, Garten W, Klenk HD, Feldmann H. Proteolytic processing of Marburg virus glycoprotein. Virology. 2000;268(1):1–6.
Volchkova VA, Klenk HD, Volchkov VE. Delta-peptide is the carboxy-terminal cleavage fragment of the nonstructural small glycoprotein sGP of Ebola virus. Virology. 1999;265(1):164–71.
Voorhees P, Deignan E, van Donselaar E, Humphrey J, Marks MS, Peters PJ, Bonifacino JS. An acidic sequence within the cytoplasmic domain of furin functions as a determinant of trans-Golgi network localization and internalization from the cell surface. EMBO J. 1995;14(20):4961–75.
Walker JA, Kawaoka Y. Importance of conserved amino acids at the cleavage site of the haemagglutinin of a virulent avian influenza A virus. J Gen Virol. 1993;74(Pt 2):311–4.
Walker JA, Molloy SS, Thomas G, Sakaguchi T, Yoshida T, Chambers TM, Kawaoka Y. Sequence specificity of furin, a proprotein-processing endoprotease, for the hemagglutinin of a virulent avian influenza virus. J Virol. 1994;68(2):1213–8.
Wang M, Shen S, Wang H, Hu Z, Becnel J, Vlak JM. Deltabaculoviruses encode a functional type I budded virus envelope fusion protein. J Gen Virol. 2017;98(4):847–52.
Webby RJ, Perez DR, Coleman JS, Guan Y, Knight JH, Govorkova EA, McClain-Moss LR, Peiris JS, Rehg JE, Tuomanen EI, Webster RG. Responsiveness to a pandemic alert: use of reverse genetics for rapid development of influenza vaccines. Lancet. 2004;363(9415):1099–103.
Weider E, Susan-Resiga D, Essalmani R, Hamelin J, Asselin MC, Nimesh S, Ashraf Y, Wycoff KL, Zhang J, Prat A, Seidah NG. Proprotein Convertase Subtilisin/Kexin Type 9 (PCSK9) single domain antibodies are potent inhibitors of low density lipoprotein receptor degradation. J Biol Chem. 2016;291(32):16659–71.
White JM, Whittaker GR. Fusion of enveloped viruses in endosomes. Traffic. 2016;17(6):593–614.
Williamson DM, Elferich J, Ramakrishnan P, Thomas G, Shinde U. The mechanism by which a propeptide-encoded pH sensor regulates spatiotemporal activation of furin. J Biol Chem. 2013;288(26):19154–65. <prodomain, His69 function as a pH-sensor>.
Wise RJ, Barr PJ, Wong PA, Kiefer MC, Brake AJ, Kaufman RJ. Expression of a human proprotein processing enzyme: correct cleavage of the von Willebrand factor precursor at a paired basic amino acid site. Proc Natl Acad Sci U S A. 1990;87(23):9378–82.
Wong E, Maretzky T, Peleg Y, Blobel CP, Sagi I. The functional maturation of A Disintegrin and Metalloproteinase (ADAM) 9, 10, and 17 requires processing at a newly identified Proprotein Convertase (PC) cleavage site. J Biol Chem. 2015;290(19):12135–46.
Wouters S, Leruth M, Decroly E, Vandenbranden M, Creemers JW, van de Loo JW, Ruysschaert JM, Courtoy PJ. Furin and proprotein convertase 7 (PC7)/lymphoma PC endogenously expressed in rat liver can be resolved into distinct post-Golgi compartments. Biochem J. 1998;336:311–6.
Xiao Y, Chen G, Richard J, Rougeau N, Li H, Seidah NG, Cohen EA. Cell-surface processing of extracellular human immunodeficiency virus type 1 Vpr by proprotein convertases. Virology. 2008;372:384–97.
Yamshchikov GV, Ritter GD, Vey M, Compans RW. Assembly of SIV virus-like particles containing envelope proteins using a baculovirus expression system. Virology. 1995;214(1):50–8.
Yang J, Goldstein JL, Hammer RE, Moon YA, Brown MS, Horton JD. Decreased lipid synthesis in livers of mice with disrupted Site-1 protease gene. Proc Natl Acad Sci U S A. 2001;98(24):13607–12.
Yang HY, Zheng NQ, Li DM, Gu L, Peng XM. Entecavir combined with furin inhibitor simultaneously reduces hepatitis B virus replication and e antigen secretion. Virol J. 2014;11:165.
Ye J, Rawson RB, Komuro R, Chen X, Dave UP, Prywes R, Brown MS, Goldstein JL. ER stress induces cleavage of membrane-bound ATF6 by the same proteases that process SREBPs. Mol Cell. 2000;6:1355–64.
Zhang X, Fugère M, Day R, Kielian M. Furin processing and proteolytic activation of Semliki Forest virus. J Virol. 2003;77(5):2981–9.
Zhou A, Martin S, Lipkind G, LaMendola J, Steiner DF. Regulatory roles of the P domain of the subtilisin-like prohormone convertases. J Biol Chem. 1998;273(18):11107–14.
Zhu X, Lindberg I. 7B2 facilitates the maturation of proPC2 in neuroendocrine cells and is required for the expression of enzymatic activity. J Cell Biol. 1995;129(6):1641–50.
Zimmer G, Budz L, Herrler G. Proteolytic activation of respiratory syncytial virus fusion protein. Cleavage at two furin consensus sequences. J Biol Chem. 2001;276(34):31642–50.
Zimmer G, Rohn M, McGregor GP, Schemann M, Conzelmann KK, Herrler G. Virokinin, a bioactive peptide of the tachykinin family, is released from the fusion protein of bovine respiratory syncytial virus. J Biol Chem. 2003;278(47):46854–61.
Author information
Authors and Affiliations
Corresponding author
Editor information
Editors and Affiliations
Rights and permissions
Copyright information
© 2018 Springer International Publishing AG, part of Springer Nature
About this chapter
Cite this chapter
Garten, W. (2018). Characterization of Proprotein Convertases and Their Involvement in Virus Propagation. In: Böttcher-Friebertshäuser, E., Garten, W., Klenk, H. (eds) Activation of Viruses by Host Proteases. Springer, Cham. https://doi.org/10.1007/978-3-319-75474-1_9
Download citation
DOI: https://doi.org/10.1007/978-3-319-75474-1_9
Published:
Publisher Name: Springer, Cham
Print ISBN: 978-3-319-75473-4
Online ISBN: 978-3-319-75474-1
eBook Packages: Biomedical and Life SciencesBiomedical and Life Sciences (R0)