Abstract
Malaria is a major worldwide health problem, specially in countries with tropical climates and remote areas. In this paper, we present an automatic system for estimating malaria parasite density in thin blood smears. The proposed approach is based on simple image processing methods that can be implemented efficiently even on low budget devices. The method has been tested on images acquired under different illumination and acquisition setups and has produced encouraging results, achieving a sensitivity of 89.3%.
You have full access to this open access chapter, Download conference paper PDF
Similar content being viewed by others
Keywords
1 Introduction
Malaria is a major worldwide public health problem. In 2014, 97 countries have been reported with continuous transmission of Malaria, and about 3.3 billion people are at risk of being contaminated [9]. In recent decades, researchers have looked for cost-effective solutions to assist health professionals in public control of epidemics and diseases, including, but not limited to, real-time diagnostic systems and epidemiological events simulations [5, 8]. Specifically, Image Processing techniques have been used been used successfully in the diagnosis of many diseases. In this article, we propose the usage of an automatic system for detecting Malaria parasites in thin blood films and computing parasite density, a fundamental information needed for a successfull treatment.
Malaria is caused by a protozoan the genus Plasmodium, where the transmission vector is a female of mosquitoes Anopheles. There are 5 species of Plasmodium can infect humans: P. falciparum, P. vivax, P. malariae, P. ovale and P. knowlesi [9].
The gold standard for malaria diagnosis, widely used around the world, is a microscopic examination of blood that uses blood films stained with Giemsa dye [7, 9]. Two techniques have been used for slide staining to red blood cells (RBC): thick blood and thin blood. The microscopy technique used in the thick blood method slide stainer is the most common and inexpensive to diagnose Malaria. The thin blood technique is used more specifically for identifying the morphology of the parasite Plasmodium spp. and in correctly identifying the species [12], and it is preferred for routine estimation because the parasites are easy to see and count in it. Accurate estimates of the number of parasites infecting, as well as the species, are very important in determining an appropriate course of treatment. If done properly, there will be an improvement in the treatment of patients and response to the drugs, especially with the P. falciparum species [7].
Here, we propose and evaluate a segmentation approach to count and identify RBCs that are healthy and infected with P. falciparum species in ring-stage. This restriction to one specie was used because of a broader access to images of thin blood films containing P. falciparum, even though the proposed method can be directly applied to identify the other species. However, a new stage of classification would be necessary to correctly identify the species.
1.1 Related Work
Recently, several papers have shown the feasibility of a detection system using image processing and artificial intelligence for medical aplications. Linder et al. [6] employed computer vision techniques to identify candidate regions based on color and size of objects to extract features using Local Binary Patterns (LBPs), local contrast and Scale-Invariant Feature Transform (SIFT) descriptors. The features extracted on the previous step are the input on linear SVM (Support Vector Machines) for classification of P. falciparum ring-stage trophozoites.
Ross et al. [13] proposed using feedforward neural networks for classification of 4 species of malaria. The neural networks are fed with image features that are extracted based on colour, texture and geometry of the objects. Another contribution is the morphological and threshold selection to identify erythrocytes (red blood cells). Pirnstill et al. [11] proposed the use of a polarized microscope, thus, exploring the properties of polarized light to identify and describe structures and properties of materials. This type of microscopy has a higher cost and requires a specific training. However, Pirnstill et al. [11] used a high fidelity and high optical resolution cell-phone based polarized light microscopy system, a much less expensive system.
Kaewkamnerd et al. [4] proposed a method divided into five phases: image acquisition; preprocessing; image segmentation; feature extraction and classification, to be used on an automatic device equiped with a motor adapted to a microscope for both detection and classification of the malaria parasite species into P. falciparum and P. vivax. In another study, Chakrabortya et al. [2] proposes an algorithm that uses morphological operations and pixel discrimination based on color to identify malaria parasites from thick smear images of P. vivax.
2 Proposed Method
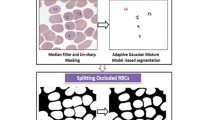
The general workflow for the proposed method is given in Fig. 1. In the remainder of this section, we will describe its major steps and talk about the reasons that impacted their development.
2.1 Image Acquisition
In this article, we deal with thin blood films, due to the fact that they allow us a better visualization of the RBCs and the morphological characteristics of the malaria parasites [12]. The data set of images was acquired and labelled by experienced parasitologists of the Microbiology Department (Drassanes Unit) of Vall d’Hebron Hospital, Barcelona, Spain, and was captured using light microscopes under an oil-immersion objective (100X) and a Nikon E5400 camera. It can be seen in Sect. 3 that, since the acquisition is performed by different specialists using different microscopes, the overall appeareance of the thin blood films change considerably over the set of images. Our method has to deal with those differences, since we want it to be of general use.
2.2 Image Preprocessing
The first steps performed in the workflow shown in Fig. 1 are part of what we call pre-processing, starting with a Gaussian blurring [3]. This reduces the noise present in the image at the cost of a small loss of signal, as we use a \(9 \times 9\) kernel to perform this operation. The resulting image is used in two other steps, a gray level conversion, whose result will be used for segmenting the RBCs, and an HSV conversion, whose result will be used to produce an HSV mask or binary image. The segmentation of the RBCs can be performed on the intensity channel alone, since the color components do not play a major role in discriminating between RBCs and the background. The Hue component of the HSV converted image will be used to detect potential plasmodia, platelets and white blood cells (WBCs), since these biological structures acquire more intense purplish colors that RBCs when stained by Giemsa [9].
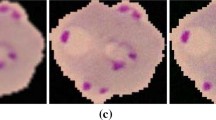
Then, an adaptive binarization is performed by using a sliding window with \(80 \times 80\) pixels. This is done because of the variations observed on the corners of some of the acquired images, where we can see different lightning and excess blurring due to change of focus. For each \(80 \times 80\) window, we compute the mean and standard deviation of the intensities and discard the window if the standard deviation is smaller that 0.03. That means that there aren’t RBCs on this window, only background, and so, the window can be discarded. The binarization of each window is performed by using the well-known nonparametric approach proposed by Otsu [10]. However, in this process, we do not take into account the intensities of pixels with high intensity stained by Giemsa, detected in the HSV mask. Figure 2 shows some results of the adaptive Otsu binarization.
We observed that some RBCs have lighter nucleus, causing the segmentation to produce cells with holes in their middle, which can impact the RBC counting step. In order to deal with this problem, we detect the outer borders of RBCs and perform a hole filling. After this step, the RBCs are sent to the connected component detection step.
2.3 Connected Component Analysis
After the holes are filled in the previous step, we analize the binary image and detect the connected components, i.e., the connected regions of pixels with the same label [14], that in this case are the contiguous regions segmented as RBCs. For each connected component we can obtain geometrical features such as area, perimeter, compactness, that can be used to estimate the total number of RBCs in an image. We empirically determined values for some of these features that allow us to estimate the number of RBCs in a single connected component. For example, we defined the minimum and maximum area thresholds for a single RBC are 500 and 1,500, respectively, while we decide if we should decompose the connected components in more than one RBC by using the value of compactness \(C = 1\), which is defined by Eq. 1:
where P is the perimeter and A is the area of the connected component.
Thus, if a connected component does not meet the area and compactness criteria defined, we try to decompose it by performing morphologic erosion [3]. An example of a connected component image can be seen on Fig. 3(f)Footnote 1. When the RBCs are conneted through a thin bridge, this process breaks the component into more than one. If the remainder of the connected component still does not meet the criteria defined for a single RBC, we then estimate the number of RBCs in the cluster of cells by dividing the area of the cluster by 1500. After the RBC cluster decomposition, the eroded RBCs are grown again by using morphologic dilation. An accurate estimation of the number of healthy RBCs is important because this is a number used to determine the parasite density, which has an impact on the treatment. The parasite density or parasitemia is considered valid only when we have a minimum counting of 500 RBCs [1], and is defined by Eq. 2:
where IRBC and TRBC are the number of infected and total RBCs, respectively.
2.4 Detecting Infected Cells
After detecting the RBCs, we use the result of the HSV mask to look for infected RBCs, as well as platelets and WBCs, since they have higher concentrations of Giemsa stain. The HSV mask is produced by looking only to the Hue channel of the HSV image and selecting the pixels with high H values. An example of such mask can be seen on Fig. 3(b)Footnote 2. According to the World Health Organization [12], on thin blood films, the parasites are located inside the RBCs. Thus, we compute the centroids of the structures detected on the HSV mask and determine on which RBC they are located by using the binary mask with the connected components. If the size of the strucure detected on the HSV mask is not within defined minimum and maximum sizes for Plasmodium trophozoites and the centroid of the structure is not inside a RBC, then it is labelled as other element type, which include WBCs, platelets and small defects. Otherwise, me mark the RBC as an infected cell.
3 Experimental Results
In order to test the efficiency of our method, we performed some experiments on an Intel Core i7-6500U (2.60 GHz 16 GB memory) computer. We used a data set containing 50 images of thin blood films infected with P. falciparum trophozoites. This data set was divided into a 20 image training data set and a 30 image testing set, and the images were labelled by an specialist. Table 1 shows the total number of cells and other structures detected by the specialist and the automated method.
Table 2 shows the results of the classification of Giemsa detected structures based on location and size. We achieved a sensitivity of 89.3% for the infected RBCs, i.e., the trophozoites, without using any shape feature. The addition of shape features for discrimination will allow us not onlt to reduce the false positive (FP) rate and even relax the rules for detecting the trophozoites so that we can also improve the true positive (TP) rate, but also to classify the infected RBCs into the different species and stages (Fig. 4).
4 Conclusion
In this paper, we proposed an automatic system for estimating malaria parasite density in thin blood smears. The proposed approach is a combination of well known image processing methods that allows us to detect and classify healthy and infected RBCs on images acquired under different illumination conditions and acquisition setups.
This is a first step towards the development of a full-fledged system that will be capable of performing classification using shape features and of identifying different stages of at least 4 species types of the Plasmodium. Moreover, we also plan to identify and classify the WBCs into its subtypes. The idea is that the system should have a small footprint, i.e., low computation and memory requirements, thus, allowing its usage on low budget devices.
Notes
- 1.
We used different colors to show different connected components on this image.
- 2.
We used red circles to label RBCs, yellow circles to label trophozoites and white circles to label other objects.
References
CDC: Diagnostic procedures (2016). https://www.cdc.gov/dpdx/diagnosticprocedures. Accessed 06 Sept 2017
Chakrabortya, K., Chattopadhyayb, A., Chakrabarti, A., Acharyad, T., Dasguptae, A.: A combined algorithm for malaria detection from thick smear blood slides. Health Med. Inform. 6(1), 179 (2015)
Gonzalez, R., Woods, R.: Digital Image Processing. Pearson/Prentice Hall, Upper Saddle River (2008)
Kaewkamnerd, S., Uthaipibull, C., Intarapanich, A., Pannarut, M., Chaotheing, S., Tongsima, S.: An automatic device for detection and classification of malaria parasite species in thick blood film. BMC Bioinform. 13(Suppl 17), S18 (2012)
Leal Neto, O.B., Cesar, A., Jones, A., Constanca, B.: The schisto track: a system for gathering and monitoring epidemiological surveys by connecting geographical information systems in real time. JMIR mHealth and uHealth 2, e10 (2014)
Linder, N., Turkki, R., Walliander, M., Mårtensson, A., Diwan, V., Rahtu, E., Pietikainen, M., Lundin, M., Lundin, J.: A malaria diagnostic tool based on computer vision screening and visualization of Plasmodium falciparum candidate areas in digitized blood smears. PLoS ONE 9(8), e104855 (2014)
Moody, A.: Rapid diagnostic tests for malaria parasites. Clin. Microbiol. Rev. 15(1), 66–78 (2002)
Oliveira, D.A., Prats, C., Espasa, M., Zarzuela Serrat, F., Montañola Sales, C., Silgado, A., Codina, L.D., Arruda, E.M., Gomez i Prat, J., Albuquerque, J.: The malaria system microapp: a new, mobile device-based tool for malaria diagnosis. JMIR Res. Protoc. 6(4), e70 (2017)
World Health Organization: World malaria report 2014. Technical report, World Health Organization (2014)
Otsu, N.: A threshold selection method from gray-level histograms. IEEE Trans. Syst. Man Cybern. 9(1), 62–66 (1979)
Pirnstill, C.W., Cote, G.L.: Malaria diagnosis using a mobile phone polarized microscope. Sci. Rep. 5, 13368 (2015)
WHO Press: Basic malaria microscopy. World Health Organization, Geneva, Switzerland (1991)
Ross, N., Pritchard, C., Rubin, D., Dusao, A.: Automated image processing method for the diagnosis and classification of malaria on thin blood smears. Med. Biol. Eng. Comput. 44(5), 427–436 (2006)
Szeliski, R.: Computer Vision: Algorithms and Applications. Texts in Computer Science. Springer, London (2010). https://doi.org/10.1007/978-1-84882-935-0
Acknowledgments
This work was supported by the Graduate Program in Systems and Computer Science (PPgSC/UFRN) and funded by CAPES-Brazil.
Author information
Authors and Affiliations
Corresponding author
Editor information
Editors and Affiliations
Rights and permissions
Copyright information
© 2018 Springer International Publishing AG, part of Springer Nature
About this paper
Cite this paper
Dantas Oliveira, A. et al. (2018). An Automatic System for Computing Malaria Parasite Density in Thin Blood Films. In: Mendoza, M., Velastín, S. (eds) Progress in Pattern Recognition, Image Analysis, Computer Vision, and Applications. CIARP 2017. Lecture Notes in Computer Science(), vol 10657. Springer, Cham. https://doi.org/10.1007/978-3-319-75193-1_23
Download citation
DOI: https://doi.org/10.1007/978-3-319-75193-1_23
Published:
Publisher Name: Springer, Cham
Print ISBN: 978-3-319-75192-4
Online ISBN: 978-3-319-75193-1
eBook Packages: Computer ScienceComputer Science (R0)