Abstract
Radiomic analysis in cancer applications enables capturing of disease-specific heterogeneity, through quantification of localized texture feature responses within and around a tumor region. Statistical descriptors of the resulting feature distribution (e.g. skewness, kurtosis) are then input to a predictive model. However, a single statistic may not fully capture the rich spatial diversity of pixel-wise radiomic expression maps. In this work, we present a new RADIomic Spatial TexturAl descripTor (RADISTAT) which attempts to (a) more completely characterize the spatial heterogeneity of a radiomic feature, and (b) capture the overall distribution heterogeneity of a radiomic feature by combining the proportion and arrangement of regions of high and low feature expression. We demonstrate the utility of RADISTAT in the context of (a) discriminating favorable from unfavorable treatment response in a cohort of N = 44 rectal cancer (RCa) patients, and (b) distinguishing short-term from long-term survivors in a cohort of N = 55 glioblastoma multiforme (GBM) patients. For both datasets, RADISTAT resulted in a significantly improved classification performance (AUC = 0.79 in the RCa cohort, AUC = 0.71 in the GBM cohort, based on randomized cross-validation) as compared to using simple statistics (mean, variance, skewness, or kurtosis) to describe radiomic co-occurrence features.
Research supported by 1U24CA199374-01, R01CA202752-01A1, R01CA208236-01A1, R21CA179327-01, R21CA195152-01, R01DK098503-02, 1 C06 RR12463-01, DOD/CDMRP PC120857, DOD/CDMRP LC130463, DOD/CDMRP W81XWH-16-1-0329, the DOD Prostate Cancer Idea Development Award; the iCorps@Ohio program, the Ohio Third Frontier Technology Validation Startup Fund, the Case Comprehensive Cancer Center Pilot Grant; VelaSano Grant from the Cleveland Clinic; the Wallace H. Coulter Foundation Program in the Department of Biomedical Engineering at CWRU. Content solely responsibility of the authors and does not necessarily represent official views of the NIH or the DOD.
You have full access to this open access chapter, Download conference paper PDF
Similar content being viewed by others
1 Introduction
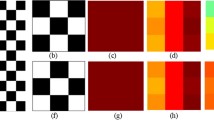
Radiomics has recently shown great promise for predicting disease aggressiveness and subtype [1]. Radiomic texture features capture pixel-wise “image texture” through quantification of local changes in image intensity values in relation to their pixel-wise arrangement within a target region of interest (ROI) [2]. Radiomics therefore may play an important role in characterizing tissue heterogeneity on radiographic imaging, based on the presence of different tissue subtypes within and around a tumor which may affect disease outcome. For example, in glioblastoma multiforme (GBM), the tumor region includes varied tissue types such as edema, necrotic core, and enhancing tumor. Similarly, in rectal cancer (RCa) patients that undergo neoadjuvant chemoradiation, treatment effects such as fibrosis and ulceration are present both within and proximal to the tumor region. As a result of such significant intra-tumoral heterogeneity, the resulting radiomic response within and around these tumors appears highly varied (see Figs. 1(a) and (f) for representative radiomic heatmaps in RCa).
Representative radiomic feature maps for 2 different rectal cancer patients, derived from post-chemoradiation MRI, with (a) poor, and (f) favorable treatment response. Note that the radiomic feature distributions significantly overlap (b), suggesting that statistical descriptors may not be able to differentiate between these patient groups. The radiomic feature is then partitioned into H (red), M (yellow), and L (cyan) expression values, shown in (c), (g), overlaid with magenta vectors to indicate connections between different expression clusters. The resulting (d), (h) textural and (e), (i) spatial phenotype that comprise RADISTAT show clear differences between the two patients.
The radiomic feature expression of a tumor ROI is commonly described using statistics of the feature distribution (e.g. skewness, kurtosis), which are then input to a machine learning classifier to make a class label prediction. While statistical descriptors may adequately describe the overall range of feature values in the tumor ROI, they may not adequately capture the spatial arrangement of differential feature expression (i.e. regions of high and low feature expression). Thus, a statistical characterization of a radiomic feature representation may not fully characterize the underlying tissue heterogeneity.
In this paper, we present a new RADIomic Spatial TexturAl descripTor (RADISTAT) to capture (a) the spatial phenotype of radiomic expression, i.e. how sub-compartments of low and high radiomic expression are spatially located relative to one another within the ROI, and (b) the textural phenotype associated with radiomic expression, i.e. whether an ROI exhibits a predominance of low or high expression sub-compartments. Figures 1(c) and (g) depict representative feature expression sub-compartments on a radiomic heatmap, based on quantizing the image into 3 expression levels (high, medium, and low).
We demonstrate the utility of RADISTAT in the context of two significant clinical problems. First, distinguishing favorable response to chemoradiation in RCA (no metastatic nodes or distant metastasis present after treatment) from poor response, via post-treatment magnetic resonance imaging (MRI). Second, differentiating long-term from short-term survivors with glioblastoma multiforme (GBM), using treatment-naive MRIs.
2 Previous Work and Novel Contributions
A few groups have recently examined alternate characterizations of radiomic features. In GBMs, an appreciation has emerged for looking at separate tumor sub-compartments, albeit using volumetric [3] or radiomic histogram [4] analysis alone. Similarly, sub-compartment-based radiomic analysis of breast MRI [5] and lung FDG PET/CT [6] have demonstrated success for predicting patient response to treatment as well as patient survival. In the work most closely related to our own [7], a gaussian mixture model of multi-parametric MR intensities was employed to define sub-compartments in GBMs. Spatial point pattern analysis was then used to perform a neighborhood analysis of these sub-compartments.
In contrast, RADISTAT leverages a more detailed radiomic characterization of tissue heterogeneity, compared to using MR intensities alone. By discretizing the rich information embedded in a radiomic heatmap into more stratified expressions, the spatial and textural relationships between the resulting radiomic “compartments” can be quantified. Sub-compartments on the radiomic feature expression map are defined through a unique 2-stage process: (1) superpixel clustering of the radiomic feature to identify spatially similar regions, and (2) re-partitioning the superpixel map to define sub-compartments based on a desired number of “expression levels” (e.g. high, medium, and low, when considering 3 expression levels). Finally, RADISTAT involves the computation of 2 distinct features: (1) the overall spatial arrangement of different sub-compartments with respect to one another, and (2) the overall proportions of different expression levels for the radiomic feature. Note that as far as we are aware, this is the first attempt at combining a spatial and proportional characterization of pixel-wise radiomic expression maps.
3 Methodology
A radiomic feature expression scene is denoted \(\mathcal {I} = (C,f)\), where C is a spatial grid of pixels c, in \(\mathbb {R}^{2}\) or \(\mathbb {R}^{3}\). Every pixel, \(c\in C\), is associated with a radiomic feature value f(c). The range of \(\mathcal {I}\) is normalized to lie between 0 and 1 (representative radiomic feature scene is visualized as a heatmap in Fig. 2). Computation of the RADISTAT descriptor comprises the following steps:
-
1.
Superpixel Clustering of Radiomic Feature Maps: Superpixel clustering of \(\mathcal {I}\) is performed using a modified version of the simple linear iterative clustering (SLIC) algorithm [8], to generate K clusters, \(\hat{C}_k \subset C\), \(k \in \{1,\dots ,K\}\). Note that in the modified SLIC implementation, K is implicitly defined based on 2 parameters: (1) the minimum number of pixels in a cluster (\(\alpha \)), and (2) the distance between initial cluster seeds (\(\beta \)). Thus for each combination of \(\alpha \) and \(\beta \), different clusterings of \(\mathcal {I}\) will be obtained. Based on superpixel clustering, \(\mathcal {I}\) is quantized to obtain a cluster map \(\hat{\mathcal {I}} = (C,g)\), where for every \(c\in \hat{C}_k\subset C\), g(c) is the average radiomic feature value within the cluster \(\hat{C}_k\). Note that \(\hat{\mathcal {I}}\) is normalized such that \(\min (g(c))=0\) and \(\max (g(c))=1\). The result of Step 1 is illustrated in Fig. 2, where the colors now represent dominant clusters of \(\hat{\mathcal {I}}\).
-
2.
Re-partitioning of Superpixel Clusters into Expression Levels: Firstly, a user-defined parameter B, which captures the desired number of expression levels, is identified. The choice of B essentially dictates how fine a variation in radiomic feature values is captured by RADISTAT. Using this input parameter B, the range of \(\hat{\mathcal {I}}\) is split into B equally spaced bins, yielding \(B+1\) thresholds \(\theta _j, j \in \{0,\dots ,B\}\). Based on the normalized range of \(\hat{\mathcal {I}}\), \(\theta _0= 0\) and \(\theta _B= 1\). These \(\theta _j\) are used to re-quantize \(\hat{\mathcal {I}}\) into an expression map, \(\tilde{\mathcal {I}} = (C,h)\), where \(\forall c\in C\), \(h(c)= \theta _j\), if \(\theta _{j-1}<g(c)<\theta _{j}\). As \(\tilde{\mathcal {I}}\) only has B unique values, any adjacent clusters which exhibit the same expression value are merged to yield M distinct partitions. A partition is defined as \(\tilde{C}_m = \{c|h(c)=\theta _j\}\), where \(m\in \{1,\dots ,M\}\), and \(\tilde{C}_m\subset C\). For ease of notation, we also define the expression value of a partition \(\tilde{C}_m\) as \(H(\tilde{C}_m) = \theta _j\), if \(\forall c\in \tilde{C}_m, h(c)=\theta _j\).
For example, when \(B=3\) (corresponding to low, medium, and high expression), the thresholds \(\theta _j=\{0,0.33,0.67,1\}\). The resulting \(\tilde{\mathcal {I}}\) will only have 3 unique values, \(\{0.33,0.67,1\}\) but can have M distinct partitions, as multiple partitions \(\tilde{C}_m\) can have the same expression value. Step 2 in Fig. 2 depicts the result of \(\tilde{\mathcal {I}}\) for \(B=3\), where each of the 3 colors represents a different expression level (low (cyan), medium (yellow), and high (red)).
-
3.
Computing the Textural Phenotype: This is obtained by quantifying the fraction of each of B expression levels in \(\tilde{\mathcal {I}}\). For \(B=3\), this means calculating what fraction of \(\tilde{\mathcal {I}}\) exhibits low, medium, or high expression. For each expression level \(\theta _j\) and \(\forall j=\{1,\dots ,B\}\),
$$\begin{aligned} {{\uptau }_{j}} = \frac{|c|h(c)=\theta _j|}{|C|}, \end{aligned}$$(1)The resulting feature is a \(1\times B\) vector \(\varvec{\uptau }=[\uptau _1,\dots ,\uptau _B]\). This is visualized via the top bar plot in Step 3 of Fig. 2.
-
4.
Computing the Spatial Phenotype: This is based on quantifying the adjacency for each pairwise combination of B expression levels in \(\tilde{\mathcal {I}}\). Considering the case of low (L), medium (M), and high (H) expression (i.e. \(B=3\)), there are 3 pairwise combinations: L-M, L-H, M-H. The adjacency of L-M is obtained by counting number of times that \(\tilde{\mathcal {I}}\) has partitions with low and medium expression adjacent to each other (similarly for L-H and M-H). For this, an adjacency graph \(G = (V,E)\) is defined, where \(V = \{v_m\}, m\in \{1,\dots ,M\}\), comprises the centroids of each of M partitions from Step 2; and \(E = \{e_{mn}\}, m,n \in \{1,\dots ,M\}\), is a set of edges. An edge in E is defined when,
$$\begin{aligned} e_{mn} = {\left\{ \begin{array}{ll} 1,&{} \text {if } \tilde{C}_m \text { adjacent to } \tilde{C}_n, m\ne n\\ 0, &{} \text {otherwise} \end{array}\right. } \end{aligned}$$(2)For every pair of expression levels \(\theta _i\) and \(\theta _j\), \(i,j\in \{1,\dots ,B\}\), the adjacency is calculated as,
$$\begin{aligned} \varsigma _{mn} = \sum e_{mn}, \text {where } H(\tilde{C}_m) = \theta _i \text { and } H(\tilde{C}_n) = \theta _j. \end{aligned}$$(3)The resulting feature is a \(1\times N\) vector \(\varvec{\varsigma } = [\varsigma _1,\dots ,\varsigma _N]\), where \(N = {B \atopwithdelims ()2}\) is the total number of expression level pairs in \(\tilde{\mathcal {I}}\). This is shown in the bottom bar plot of Step 3 in Fig. 2.
-
5.
Constructing RADISTAT descriptor: RADISTAT is constructed by concatenating \(\varvec{\varsigma }\) and \(\varvec{\uptau }\) to yield a \(1\times (B+N)\) vector.
4 Experimental Design
4.1 Data Description
Dataset 1: A retrospective cohort of 44 RCa patients who underwent neoadjuvant chemoradiation were imaged with 3 Tesla T2-w MRI prior to rectal excision. Patients were histologically classified as favorable response to treatment (\(n=32\)) or poor (\(n=12\)) response. Favorable response in the context of rectal cancer is defined as the absence of residual disease from metastasizing or spreading to surrounding lymph nodes, which is extremely difficult to determine on visual inspection of the MRI.
Dataset 2: A cohort of 55 patients with GBM were initially diagnosed using Gadolinium-contrast (Gd-c) T1-w MRI and were studied retrospectively for time to overall survival (OS). Following standard treatment regimes (chemoradiation or surgery), 26 patients were reported to have long-term survival (\(OS>540\) days) and 29 patients reported short-term survival (\(OS<240\) days).
4.2 Implementation Details
Figure 2 demonstrates the workflow of RADISTAT, and its implementation in the context of clinical problems in RCa and GBM. For each dataset considered, a representative 2D section was obtained from the middle of isotropically resampled volumes, and the region of interest was annotated by an expert radiologist. 12 gray level co-occurrence matrix (GLCM) features were extracted on a pixel-wise basis [9] from every 2D section. These were entropy, energy, inertia, correlation, information measures 1 and 2, sum and difference averages, variances, and entropies. The number of feature expression levels was fixed at \(B=3\), corresponding to high, medium, and low radiomic expression levels.
Parameter sensitivity was evaluated for different combinations of superpixel parameters (\(\alpha \in \{3,5,7\}\), \(\beta \in \{5,10,15,20\}\)) and all 12 GLCM features (240 combinations). Each combination was evaluated using a linear discriminant analysis (LDA) classifier, in order to differentiate the 2 patient groups in each cohort. Classifier performance was evaluated using 25 runs of 3-fold cross-validation. Optimal combination of superpixel parameters for each feature was selected based on averaging the area under the receiver-operator curve (AUC) across all 25 runs. \(\alpha =7\) and \(\beta =5\) were empirically found to be optimal parameters based on highest AUC for each problem and were used for further evaluation.
We compared RADISTAT against 4 statistical descriptors (mean, variance, skewness, and kurtosis of the radiomic expression distribution), \(\varvec{\uptau }\), and \(\varvec{\varsigma }\). Kruskal-Wallis multiple comparison testing was performed to determine statistical significance, based on adjusted p-values via the Bonferroni correction.
5 Results
5.1 Distinguishing Treatment Response in Rectal Cancer
Figure 1 shows a representative low and high clinically staged patient for rectal cancer following chemoradiation treatment. The heatmaps shown in the first column depict the radiomic feature representation of a single GLCM descriptor, correlation, for each pixel, where higher values of correlation are shown in red while lower values are shown in blue. Distributions of the radiomic feature expression between the two patients are shown in the second column. While there appears to be minimal separation in the distribution curves of the radiomic expression between the two pathologic responses (second column), re-quantizing the radiomic heatmap through superpixel clustering and partitioning (third column) reveals underlying differences in the frequency of binned expression levels (\(\varvec{\uptau }\), fourth column) and spatial arrangement of the expression clusters (\(\varvec{\varsigma }\), fifth column). The magenta vectors overlaid on the partitioned radiomic expression level map in column 3 indicate the presence of an adjacent edge between two different expression level clusters. It is interesting to note that the patient with favorable response has a higher proportion of medium to high expression and more graph connections with high expression clusters than the patient with poor response. The corresponding quantitative results in Fig. 4(a) demonstrate that RADISTAT significantly outperformed top-ranked statistics for the three highest performing GLCM features energy, correlation, and difference average (\(\text {p}<0.001\) for each). RADISTAT typically performed higher than \(\varvec{\uptau }\) and was always comparable with \(\varvec{\varsigma }\). We also considered the use of a \(1\times 4\) vector of statistical descriptors, but this always resulted in a worse AUC than the 3 best performing statistics (not shown).
Representative radiomic feature heatmaps for (a) long-term GBM survivor, and (f) short-term GBM survivor. The feature distributions in (b) show significant overlap. When considering (c), (g) \(B=3\) expression levels for RADISTAT, histograms of the proportion of expression levels and adjacent connections between different expression levels reveal underlying differences in the (d), (h) textural and (e), (i) spatial phenotypes between the two survival outcomes.
5.2 Predicting Overall Survival in GBMs
Figure 3 shows representative results for GBM patients with long-term survival (top row) and short-term survival (bottom row). Radiomic heatmaps and expression maps shown are for the GLCM feature inertia, which is a measure of contrast within a neighborhood of pixels. A markedly greater proportion of medium expression as well as more graph connections between low and high expression compartments was observed in short-term survival GBM patients. RADISTAT quantitatively outperformed the best statistic and \(\varvec{\uptau }\) for the highest performing GLCM features inertia (\(\text {p}<0.001\)), information measure 1 (\(\text {p}<0.001\)), and difference variance (\(\text {p}<0.001\)); while achieving marginally higher AUCs than \(\varvec{\varsigma }\) alone (see Fig. 4(b)).
Average AUCs across 25 runs of 3-fold cross validation for (a) rectal cancer dataset, and (b) GBM dataset, for top 3 GLCM features in each experiment. RADISTAT (red bars) resulted in a consistently higher performance than any compared strategy: the best performing statistical descriptor (magenta bars), individual textural (green) and spatial (blue) components of RADISTAT.
6 Concluding Remarks
In this work, we presented a novel radiomic descriptor, RADISTAT, which ascribes a combined textural and spatial phenotype to a radiomic feature expression map to better characterize tissue heterogeneity. In a cross-validation setting, RADISTAT was found to significantly outperform commonly applied statistical measures for representing the radiomic expression map in the context of (a) distinguishing favorable from poor treatment response for RCa patients, and (b) predicting survival in GBM patients. In future work, we will seek to understand the correlation of RADISTAT with specific pathological phenotypes, extend its implementation to 3D, as well as apply it across other disease sites.
References
Aerts, H., et al.: Decoding tumour phenotype by noninvasive imaging using a quantitative radiomics approach. Nat. Commun. 5, 4006 (2014)
Antunes, J., et al.: Radiomics analysis on FLT-PET/MRI for characterization of early treatment response in renal cell carcinoma: a proof-of-concept study. Transl. Oncol. 9(2), 155–162 (2016)
Zhou, M., et al.: Radiologically defined ecological dynamics and clinical outcomes in glioblastoma multiforme: preliminary results. Transl. Oncol. 7(1), 5–13 (2014)
Zhou, M., et al.: Identifying spatial imaging biomarkers of glioblastoma multiforme for survival group prediction. J. Magn. Res. Imaging 46(1), 115–123 (2016)
Wu, J., et al.: Intratumor partitioning and texture analysis of dynamic contrast-enhanced (DCE)-MRI identifies relevant tumor subregions to predict pathological response of breast cancer to neoadjuvant chemotherapy. J. Magn. Reson. Imaging 44(5), 1107–1115 (2016)
Wu, J., et al.: Robust intratumor partitioning to identify high-risk subregions in lung cancer: a pilot study. Int. J. Radiat. Oncol. Biol. Phys. 95(5), 1504–1512 (2016)
Lee, J., et al.: Spatial habitat features derived from multiparametric magnetic resonance imaging data are associated with molecular subtype and 12-month survival status in glioblastoma multiforme. PLoS ONE 10(9), e0136557 (2015)
Achanta, R., et al.: SLIC superpixels compared to state-of-the-art superpixel methods. IEEE Trans. Pattern Anal. Mach. Intell. 34(11), 2274–2282 (2012)
Haralick, R., et al.: Textural features for image classification. IEEE Trans. Syst. Man Cybern. SMC–3(6), 610–621 (1973)
Author information
Authors and Affiliations
Corresponding author
Editor information
Editors and Affiliations
Rights and permissions
Copyright information
© 2017 Springer International Publishing AG
About this paper
Cite this paper
Antunes, J., Prasanna, P., Madabhushi, A., Tiwari, P., Viswanath, S. (2017). RADIomic Spatial TexturAl descripTor (RADISTAT): Characterizing Intra-tumoral Heterogeneity for Response and Outcome Prediction. In: Descoteaux, M., Maier-Hein, L., Franz, A., Jannin, P., Collins, D., Duchesne, S. (eds) Medical Image Computing and Computer-Assisted Intervention − MICCAI 2017. MICCAI 2017. Lecture Notes in Computer Science(), vol 10434. Springer, Cham. https://doi.org/10.1007/978-3-319-66185-8_53
Download citation
DOI: https://doi.org/10.1007/978-3-319-66185-8_53
Published:
Publisher Name: Springer, Cham
Print ISBN: 978-3-319-66184-1
Online ISBN: 978-3-319-66185-8
eBook Packages: Computer ScienceComputer Science (R0)