Abstract
Intra-operative measurements of tissue shape and multi/hyperspectral information have the potential to provide surgical guidance and decision making support. We report an optical probe based system to combine sparse hyperspectral measurements and spectrally-encoded structured lighting (SL) for surface measurements. The system provides informative signals for navigation with a surgical interface. By rapidly switching between SL and white light (WL) modes, SL information is combined with structure-from-motion (SfM) from white light images, based on SURF feature detection and Lucas-Kanade (LK) optical flow to provide quasi-dense surface shape reconstruction with known scale in real-time. Furthermore, “super-spectral-resolution” was realized, whereby the RGB images and sparse hyperspectral data were integrated to recover dense pixel-level hyperspectral stacks, by using convolutional neural networks to upscale the wavelength dimension. Validation and demonstration of this system is reported on ex vivo/in vivo animal/human experiments.
You have full access to this open access chapter, Download conference paper PDF
Similar content being viewed by others
Keywords
These keywords were added by machine and not by the authors. This process is experimental and the keywords may be updated as the learning algorithm improves.
1 Introduction
Using intra-operative information to aid surgical planning, navigation, and decision making is important for minimally invasive and robotic surgery. The data is mainly collected via endoscopes and other integrated hardware to provide real-time texture and color information. By data analysis the tissue surface shape can be extracted to register the intra- and pre-operative information from imaging modalities like CT and MRI [1]. Intra-operative optical modalities such as multi/hyperspectral imaging (MSI/HSI) also have significant clinical impact, e.g. (1) narrow band imaging for vascular visualization; (2) oxygen saturation for intra-operative perfusion monitoring and clinical decision making; (3) tissue classification and pathology identification [2, 3].
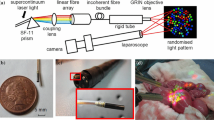
Previously, a SL-enabled 3D tissue surface shape and hyperspectral imaging system was presented [4]. This used an optical fiber bundle with the fibers arranged in linear and circular arrays, respectively, at either end (Fig. 1(a)). In SL mode dispersed supercontinuum laser light was directed onto the linear array to emerge from the circular array as a spectrally encoded spot pattern. The tissue shape could be reconstructed sparsely if this light pattern was projected onto the surface and analyzed. Unlike passive stereo techniques, this SL reconstruction is not restricted by the texture information on the object surface. In HSI mode, the endoscopic white light illuminated the target surface. The reflected light was captured by the circular fiber array, emerged from the linear array, and imaged onto a slit HSI camera. The positions of detected spots in SL mode therefore indicated the locations of HSI signal on the white light images. However, the system did not provide surgeons with a WL view containing texture information, since no RGB images were captured. Furthermore, it also suffered from sparse reconstructed surface and HSI signal, due to the finite number of fibers in the bundle. In this work, these two problems were addressed.
In this work a chopper wheel was used to stroboscopically switch between the SL and WL modes, to provide the surgeons with WL view. Both views were utilized to provide a quasi-dense reconstruction with known scale, using both SL and SfM [5]. This procedure was applied on a GPU to guarantee fast processing. Secondly, a deep learning-based method was studied to generate pixel-level dense multispectral image (MSI) stacks from captured RGB images and the sparse HSI signals.
HSI systems can be divided into two main types: spatial (e.g. with slit HSI camera) and spectral scanning systems (e.g. liquid crystal tunable filter (LCTF), or filter wheel multispectral cameras). However, there is always a trade-off between spatial resolution, spectral resolution and acquisition time, which affects surgical applications where the tissue is deformable and moving. Du et al. proposed to use non-rigid registration to align mismatched HSI stacks [6], but such methods are limited to relatively long, off-line processing. Recently, convolutional neural networks have been used to solve the image super-resolution problem, mainly to increase the spatial resolution of input images. Shi et al. proposed fully convolutional networks to improve the image quality [7]; Oktay et al. applied residual networks to upscale low-resolution 3D MRI data [8]. In this paper, we developed a model to merge information from dense RGB images and sparse HSI signals to predict the corresponding dense 3D MSI stack with 24 channels. This was realized by upscaling the RGB images on its channel dimension and integrating sparse HSI signals to fine-tune the spectral shape on different locations. We refer to this method as “super-spectral-resolution”, i.e., achieving spatial super-resolution of sparse multispectral measurements by upscaling dense WL images in the spectral domain. The proposed model was trained on in vivo animal tissues, and tested on both ex vivo and in vivo in human and animal experiments.
In a nutshell, there are three key contributions in this work: (1) hardware improvement to provide WL views; (2) combination of SL and SfM for quasi-dense reconstruction; (3) real-time dense MSI using RGB images and sparse HSI signals.
2 Materials and Methods
2.1 Interleaved SL and WL Views
Rapid stroboscopic switching between WL and SL was achieved using an optical chopper wheel (3501 Optical Chopper; New Focus, Inc., USA) [9] as shown in Fig. 1(a). Two fibre optic light cables were used for WL: one was connected to a xenon lamp and the other to the laparoscope. Their free ends were then positioned against each other, separated by a 2 mm air gap through which the chopper wheel could pass. The chopper was mounted so that the output of the supercontinuum laser also passed through the wheel, and the SL and WL beam paths were alternately blocked or transmitted as it turned. The result was that the light emerging from the tip of the instrument switched between SL and xenon at the chopping frequency. Separately, a computer-controlled signal generation device (NI USB-6211; National Instruments Corporation, USA) was used to produce two synchronized square waveforms of variable frequency and phase. One was used to trigger image acquisition by the CCD camera, while the other controlled the rotation frequency and the phase of the chopper wheel. The trigger frequency was set to twice that of the chopper and the phase adjusted so that the acquired frames comprised of alternating SL and WL-illuminated images.
A tip adapter was 3D printed to mount the SL probe on a rigid endoscope (5 mm diameter Hopkins II Optik 30o, Karl Storz GmbH, Germany). This adapter was cylindrical (12 mm diameter), with two channels to house the endoscope and SL probe. The angle and baseline of these two channels were set to 10° and 5 mm, to maximize triangulation accuracy for surface reconstruction within ~1.5–4 cm working distances.
2.2 Tissue Surface Feature Tracking and Shape Measurement
In this work, we propose to combine information from both the SL and WL images. Previously, an SL reconstruction technique with fully convolutional networks (FCN) has been proposed, and worked robustly at a frame rate of ~12 FPS [4]. However, due to in vivo factors like strong light tissue interaction and CCD over-exposure, using SL alone does not always return a dense reconstruction. Therefore, we combined SL and SfM on a GPU to increase the reconstruction density and robustness (Fig. 1(b)).
Surface Reconstruction Using Monocular SfM.
In this work, a method combining SURF-based feature detection and LK optical flow-based tracking has been proposed to perform a correspondence search. Several criteria were applied to exclude the tracking outliers, including the feature descriptor difference, flow vector length, temporal smoothness, symmetric optical flow, and RANSAC in essential matrix estimation. For the surface reconstruction we assumed the surface was rigid in a small time window. Then the relative position between the cameras in the two frames, as well as the up-to-scale 3D positions of the feature points, can be estimated using singular value decomposition (SVD) and examining all four possible solutions. Given enough correspondences, the surface can be measured using two adjacent frames.
Combination of Reconstruction Results from SL and SfM.
Since each WL image had two temporally adjacent SL images, the average shape reconstructed from two SL frames was used to register the SfM reconstruction results with scale information.
2.3 Super-Spectral-Resolution Imaging
Super-resolution, recovering high-resolution (HR) images from their low resolution (LR) counterparts, is an ill-posed method, where one LR input could be mapped to multiple HR outputs. To solve this problem, two assumptions were made in this work: (1) The HR information is redundant in the HR images and could be partially extracted from the LR ones. (2) The mapping from LR to HR can be learnt from training sets containing data similar to the unseen data. The proposed approach upscaled the spectral dimension rather than the spatial dimension. Two models were developed: one recovers MSI stacks from RGB images only, while the other combines RGB images with the sparsely collected hyperspectral signal to further refine the MSI prediction.
Model 1 - Recovering MSI Stacks from RGB Images.
An RGB image was considered as an MSI stack with 3 spectral channels. The proposed model (Fig. 2(a)) looked for a mapping from an \( M \times N \times 3 \) stack to an \( M \times N \times 24 \) MSI stack, where \( M \times N \) stands for the image spatial resolution. This model consists of two main stages:
-
Upscaling the input in the spectral dimension. Four 3D transposed convolutional layers were piled together to transform the input from \( M \times N \times 3 \) to \( M \times N \times 24 \).
-
High spectral frequency signal extraction. This extracts and combines the high frequency signal with LR stacks. This stage was implemented using a residual block which introduces a “shortcut” to reduce the degradation of the training accuracy problem when deep networks are used. In our model the convolutional mapping \( F\left( x \right) \) was used to extract the high frequency from input \( x \), and then added to the “shortcut” \( x \) input itself which represents high frequency content.
The structure of model 1 can be found in Fig. 2(a). This mapping achieved generally good spectral prediction but still with noticeable errors. To refine the predication, we extended model 1 to incorporate spatially sparse HSI signals captured using the system’s HSI mode.
Model 2 - Recovering HSI Stacks from RGB Images and Sparse Spectral Signals.
RGB images provided high spatial but low spectral resolution; while HSI mode had low spatial but high spectral resolution. Due to the sparsity of the hyperspectral signal, RGB was used as the main contributor for MSI stack estimation, then the HSI signal was applied to refine the estimation.
Model 2 takes three inputs: an RGB image (\( M \times N \times 3 \)), a density map (\( M \times N \)) indicating the locations where the HSI is collected, and a sparse stack (\( M \times N \times 24 \)) containing the sparse HSI signal. Model 2 added a “merging stage” on top of Model 1 (Fig. 2(b)), where all inputs were integrated. The HSI data was concatenated with the element-wise product between the density map and the HSI stack recovered from RGB. The final spatially dense MSI stack was estimated following a convolution.
Training and Prediction.
Choosing a training set that provides adequate prior knowledge is of great importance for accurate MSI recovery. In this work, MSI stacks collected in vivo during animal trials have been used for training and testing. The stacks (\( H \)) were collected using an LCTF endoscopic imager [3], and different spectra were registered to create spectrally matched stacks. The transmission spectrum (\( h \)) for an RGB camera (Thorlabs DCU223C) was utilized to generate the synthetic RGB images (\( R \)) from HSI stacks, with \( R = h \otimes H \). The density map (\( D_{hsi} \)) for the sparse HSI signal was produced using previous spot segmentation results, with Gaussian distribution (max = 1) filled at each spot location (where the sparse spectral signal comes from). The density map for the RGB image (\( D_{rg} \)) was defined by \( D_{rgb} = 1 - D_{hsi} \). The sparse HSI stack (\( H_{s} \)) was the element-wise product between the density map and the HSI stack (\( H_{s} = D_{hsi} . *H \)).
To guarantee sufficient training samples, model 1 was trained on individual pixel spectral vectors instead of whole MSI stacks. In this case convolutions were applied along the spectral dimension, so that the trained network can be applied to inputs with arbitrary spatial dimensions. When training model 2, the network was initialized by the trained parameters from model 1. A two-stage training strategy was adopted instead of training directly from scratch: the parameters in the shared layers with model 1 were frozen while the “mergence” layers were updated; then all the parameters were updated until convergence. Both models were trained using Adam optimizer and L2-norm loss function. In prediction, RGB images were captured by the same camera, and the sparse HSI signal came from a HSI camera. Training and prediction were implemented using Tensorflow [10]. The prediction costs ~120 ms per frame on a PC (OS: Ubuntu 14.04; CPU: i7-3770; GPU: NVIDA GTX TITAN X).
3 Experimental Results
In vivo animal experimental data (MSI stack from 50 pig bowel, 21 rabbit uterus, 10 sheep uterus) were used to train and validate both models. By mixing the data from different sources and data augmentation, a 5-fold leave one-out cross-validation (LOOCV) was applied on a dataset containing 243 MSI stacks; each fold contained 200 MSI stacks for training and the remaining for testing.
Given the ground truth the peak signal-to-noise ratio (\( {{PSNR = 20}}log_{10} ( 2 5 5 /\sqrt {{MSE}} ) \), MSE: mean square error) was adopted to evaluate the performances of both models. In the validation on average model 2 demonstrated significantly higher PSNR (~30.4) compared with model 1 (~28.5). The average PSNR on different wavelengths are shown in Fig. 3(b). In order to intuitively show the difference between two models, the estimated multispectral signals from 5 points, randomly chosen from representative areas in one pig bowel image, are compared (Fig. 3(a)). Although model 1 provided an estimation that generally fitted the ground truth, it suffered from large errors at some wavelengths. To the contrary model 2 provided improved accuracy over the entire spectral range. The pixel level PSNR maps for the estimated MSI regarding the same image are shown in Fig. 3(c, d). Excluding the saturation area, the minimum and mean PSNR are 13.1 and 34.7 for model 1, and 14.1 and 42.0 for model2.
Evaluation of transfer learning results is of great importance on machine learning problems, especially the clinical ones, where high model generalization capability is required. Thus, we also trained our models on data from different sources and tested them on each other. Table 1 lists the transfer learning results on two models, showing the importance of integrating the sparse HSI signal for more accurate MSI estimation.
Acquiring the MSI stack is a prerequisite for imaging modalities like oxygen saturation and narrow band imaging, which could provide information to aid diagnosis and surgical navigation. As image examples, the synthetic oxygen saturation and narrow band images, estimated from the predicted MSI stacks, are overlaid onto the 3D reconstructed surfaces from ex vivo and in vivo human experiments (Fig. 4).
4 Discussion and Conclusion
We have proposed a system capable of reconstructing tissue surface shape and recovering dense multispectral signals. The implementation of interleaved SL and WL imaging provided WL views with shape and texture information that could be extracted for further applications e.g., object tracking or visual servoing, to benefit MIS and robotic surgery. The SfM pipeline can be further updated to state-of-the-art algorithms in future work. A near real-time (~8 FPS) algorithm has been proposed to recover dense pixel-level multispectral signals. The accuracy and robustness of this algorithm have been demonstrated statistically and intuitively using different experimental results, in vivo and ex vivo from animal and human studies. Different imaging modalities derived from the estimated MSI stack, such as oxygen saturation and narrow band imaging, were shown. The performance should be validated by further experiments on human tissue, especially abnormal structures like tumors and polyps. We believe the “super-spectral-resolution” algorithm can also benefit other general HSI acquisition modalities to greatly reduce scanning time with little compromise in performance.
References
Maier-Hein, L., Mountney, P., Bartoli, A., Elhawary, H., Elson, D., Groch, A., Kolb, A., Rodrigues, M., Sorger, J., Speidel, S., Stoyanov, D.: Optical techniques for 3D surface reconstruction in computer-assisted laparoscopic surgery. Med. Image Anal. 17, 974–996 (2013)
Lu, G., Fei, B.: Medical hyperspectral imaging: a review. J. Biomed. Opt. 19(1), 010901 (2014)
Clancy, N.T., Arya, S., Stoyanov, D., Singh, M., Hanna, G.B., Elson, D.S.: Intraoperative measurement of bowel oxygen saturation using a multispectral imaging laparoscope. Biomed. Opt. Exp. 6, 4179–4190 (2015)
Lin, J., Clancy, N.T., Sun, X., Qi, J., Janatka, M., Stoyanov, D., Elson, D.S.: Probe-based rapid hybrid hyperspectral and tissue surface imaging aided by fully convolutional networks. In: Ourselin, S., Joskowicz, L., Sabuncu, M.R., Unal, G., Wells, W. (eds.) MICCAI 2016. LNCS, vol. 9902, pp. 414–422. Springer, Cham (2016). doi:10.1007/978-3-319-46726-9_48
Hartley, R., Zisserman, A.: Multiple View Geometry in Computer Vision. Cambridge University Press, Cambridge (2003)
Du, X., Clancy, N., Arya, S., Hanna, G.B., Kelly, J., Elson, D.S., Stoyanov, D.: Robust surface tracking combining features, intensity and illumination compensation. Int. J. Comput. Assist. Radiol. Surg. 10, 1915–1926 (2015)
Shi, W., Caballero, J., Huszár, F., Totz, J., Aitken, A.P., Bishop, R., Rueckert, D., Wang, Z.: Real-time single image and video super-resolution using an efficient sub-pixel convolutional neural network. In: 2016 IEEE Conference on Computer Vision and Pattern Recognition (CVPR), pp. 1874–1883 (2016)
Oktay, O., et al.: Multi-input cardiac image super-resolution using convolutional neural networks. In: Ourselin, S., Joskowicz, L., Sabuncu, M.R., Unal, G., Wells, W. (eds.) MICCAI 2016. LNCS, vol. 9902, pp. 246–254. Springer, Cham (2016). doi:10.1007/978-3-319-46726-9_29
Clancy, N.T., Stoyanov, D., Yang, G.-Z., Elson, D.S.: Stroboscopic illumination scheme for seamless 3D endoscopy, p. 82140M-82146 (2012)
Abadi, M., Barham, P., Chen, J., Chen, Z., Davis, A., Dean, J., Devin, M., Ghemawat, S., Irving, G., Isard, M., Kudlur, M., Levenberg, J., Monga, R., Moore, S., Murray, D.G., Steiner, B., Tucker, P., Vasudevan, V., Warden, P., Wicke, M., Yu, Y., Zheng, X.: TensorFlow: a system for large-scale machine learning. ArXiv e-prints 1605 (2016)
Ethics Statement
The ethics approval for human study was covered by Central London Research Ethics Committee (reference No. 10/H0718/55), animal study was conducted under UK Home Office license (reference No. 70/24843, 70/7508, 70/6927, 8012639).
Author information
Authors and Affiliations
Corresponding author
Editor information
Editors and Affiliations
Rights and permissions
Copyright information
© 2017 Springer International Publishing AG
About this paper
Cite this paper
Lin, J. et al. (2017). Endoscopic Depth Measurement and Super-Spectral-Resolution Imaging. In: Descoteaux, M., Maier-Hein, L., Franz, A., Jannin, P., Collins, D., Duchesne, S. (eds) Medical Image Computing and Computer-Assisted Intervention − MICCAI 2017. MICCAI 2017. Lecture Notes in Computer Science(), vol 10434. Springer, Cham. https://doi.org/10.1007/978-3-319-66185-8_5
Download citation
DOI: https://doi.org/10.1007/978-3-319-66185-8_5
Published:
Publisher Name: Springer, Cham
Print ISBN: 978-3-319-66184-1
Online ISBN: 978-3-319-66185-8
eBook Packages: Computer ScienceComputer Science (R0)