Abstract
New advances in genome engineering technologies, such as efficient programmable CRISPR nucleases, have enabled new advances in forward and reverse genetic studies. Here, I discuss recent work from our group combining top-down approaches like genome-wide loss-of-function screens and bottom-up approaches like disease variant modeling in human stem cells and stem cell-derived cortical neurons.
You have full access to this open access chapter, Download chapter PDF
Similar content being viewed by others
Introduction
Patient-sequencing studies have yielded large lists of disease-associated gene variants but it has been difficult to establish a causal role based solely on genetics. New methods are needed for rapidly understanding the effects of these variants and ascertaining whether the variants directly influence disease-related phenotypes. At the IPSEN meeting “Genome Editing in Neurosciences,” I presented two approaches (top-down and bottom-up) for harnessing new genome engineering techniques to decipher the roles of genetic variants in human health and disease.
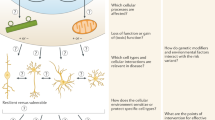
Top-down approaches utilize large-scale pooled libraries of genome-engineering reagents to start with a large, minimally biased hypothesis space and identify relevant variants via a single phenotypic selection (Fig. 1, right). In contrast, bottom-up approaches start with a handful of genetic variants nominated by strong genetic data, e.g., genome-wide association studies, case-control, family linkage studies, etc., and they examine a wide range of phenotypes (Fig. 1, left).
Schematic diagram of the dynamic interplay between top-down and bottom-up genetic approaches. Top-down approaches (right) identify new gene candidates that can shape/reduce the space of hypotheses for more detailed bottom-up (left) cellular models and phenotyping. Top-down approaches are unbiased or minimally biased to cast a wide net of possible genetic hypotheses and use phenotypic selection to identify putative disease-associated gene variants. Bottom-up approaches focus on a smaller set of variants but usually provide a more detailed phenotypic analysis of different molecular/cellular/circuit aspects of each genetic variant. Candidate variants for the bottom-up approach can be derived from either genetic evidence (e.g., patient sequencing studies) or from top-down approaches like genome-wide CRISPR screens
Top-Down Approaches Using Genome-Wide CRISPR Screens
The microbial CRISPR-Cas9 nuclease from S. pyogenes can be guided to specific DNA sequences using a 20 bp guide sequence. Given the short length of the guide sequence, we have been able to use oligonucleotide array synthesis techniques to create libraries of thousands of guide sequences in a pooled format. By designing pooled libraries to target all genes in a specific genome, we have created a new tool for functional genomic screens (Sanjana 2016). Genome-scale CRISPR knock-out (GeCKO) screens use the consistent phenotypic enrichment of multiple CRISPR reagents targeting the same gene to lend evidence to the gene’s role in a particular disease. Using a GeCKO library targeting ~18,000 genes with 64,751 guide sequences, we have found loss-of-function mutations that confer resistance to the BRAF inhibitor vemurafenib in human melanoma cells (Shalem et al. 2014). Using a second-generation GeCKO library in a mouse model, we performed an in vivo screen to identify driver mutations that trigger metastasis to the lung (Sanjana et al. 2014; Chen et al. 2015). Recently, we have expanded the scope of CRISPR pooled screens to also include noncoding regions of the genome (Sanjana et al. 2016; Wright and Sanjana 2016).
Bottom-Up Approaches Using Exome Sequencing in Autism
Whole exome and whole genome sequencing have ushered in a revolution in identifying rare, disease-associated variants. Several whole exome sequencing studies have pinpointed rare de novo variants associated with autism spectrum disorder by examining exomes from autistic individuals and comparing them to parental exomes (Iossifov et al. 2012; Neale et al. 2012; O’Roak et al. 2012; Sanders et al. 2012). The next logical step is to create relevant cellular models to better understand the mechanisms through which these variants work and to serve as a platform for drug screens and therapeutic testing. There are two major roadblocks for building these kinds of cellular models. The first one concerns gene editing and, over the past 3 years, has become largely historical: Until recently, genome engineering in human stem cells and neurons has been challenging but transfection of CRISPR plasmids or ribonucleoproteins provides an easy, efficient technique for engineering human cells (Peters et al. 2008; Swiech et al. 2015). The second major hurdle has been neural differentiation. Common protocols to differentiate neurons from human stem cells, such as dual SMAD inhibition or embryoid body differentiation, require months to create mature neurons (Zhang et al. 2001; Chambers et al. 2009). Recently, we and others have demonstrated that viral overexpression of Neurogenin 1 or Neurogenin 2 can rapidly drive stem cells into a homogeneous culture of mature cortical neurons (Zhang et al. 2013; Busskamp et al. 2014). These neurons display robust electrophysiological activity within just 2–3 weeks after the start of differentiation, making them ideally suited for synaptic assays, calcium imaging and neurophysiology. We are now moving forward with phenotypic analyses of de novo mutations in autism using the combined CRISPR-Neurogenin platform for rapid mutagenesis and human neuron profiling.
Taken together, these new technologies in genome engineering—enabled in large part by CRISPR nucleases and related transformative methods—have improved our ability to perform forward and reverse genetic assays in relevant model systems. A major challenge for neuroscience is finding clear phenotypes that accurately reflect complex diseases, such as schizophrenia or autism. Despite these challenges, a combination of top-down and bottom-up approaches will pave the way for a clearer understanding of the human brain in healthy and disease states.
References
Busskamp V, Lewis NE, Guye P, Ng AHM, Shipman SL, Byrne SM, Sanjana NE, Murn J, Li Y, Li S, Stadler M, Weiss R, Church GM (2014) Rapid neurogenesis through transcriptional activation in human stem cells. Mol Syst Biol 10:760
Chambers SM, Fasano CA, Papapetrou EP, Tomishima M, Sadelain M, Studer L (2009) Highly efficient neural conversion of human ES and iPS cells by dual inhibition of SMAD signaling. Nat Biotechnol 27:275–280
Chen S, Sanjana NE, Zheng K, Shalem O, Lee K, Shi X, Scott DA, Song J, Pan JQ, Weissleder R, Lee H, Zhang F, Sharp PA (2015) Genome-wide CRISPR screen in a mouse model of tumor growth and metastasis. Cell 160:1246–1260
Iossifov I, Ronemus M, Levy D, Wang Z, Hakker I, Rosenbaum J, Yamrom B, Lee YH, Narzisi G, Leotta A, Kendall J, Grabowska E, Ma B, Marks S, Rodgers L, Stepansky A, Troge J, Andrews P, Bekritsky M, Pradhan K, Ghiban E, Kramer M, Parla J, Demeter R, Fulton LL, Fulton RS, Magrini VJ, Ye K, Darnell JC, Darnell RB, Mardis ER, Wilson RK, Schatz MC, McCombie WR, Wigler M (2012) De novo gene disruptions in children on the autistic spectrum. Neuron 74:285–299
Neale BM, Liu L, Ma’ayan A, Samocha KE, Sabo A, Lin CF, Stevens C, Wang LS, Makarov V, Polak P, Yoon S, Maguire J, Crawford EL, Campbell NG, Geller ET, Valladares O, Schafer C, Liu H, Zhao T, Cai G, Lihm J, Dannenfelser R, Jabado O, Peralta Z, Nagaswamy U, Muzny D, Reid JG, Newsham I, Wu Y, Lewis L, Han Y, Voight BF, Lim E, Rossin E, Kirby A, Flannick J, Fromer M, Shakir K, Fennell T, Garimella K, Banks E, Poplin R, Gabriel S, DePristo M, Wimbish JR, Boone BE, Levy SE, Betancur C, Sunyaev S, Boerwinkle E, Buxbaum JD, Cook EH Jr, Devlin B, Gibbs RA, Roeder K, Schellenberg GD, Sutcliffe JS, Daly MJ (2012) Patterns and rates of exonic de novo mutations in autism spectrum disorders. Nature 485:242–245
O’Roak BJ, Vives L, Girirajan S, Karakoc E, Krumm N, Coe BP, Levy R, Ko A, Lee C, Smith JD, Turner EH, Stanaway IB, Vernot B, Malig M, Baker C, Reilly B, Akey JM, Borenstein E, Rieder MJ, Nickerson DA, Bernier R, Shendure J, Eichler EE (2012) Sporadic autism exomes reveal a highly interconnected protein network of de novo mutations. Nature 485:246–250
Peters DT, Cowan CA, Musunuru K (2008) Genome editing in human pluripotent stem cells. Harvard Stem Cell Institute, Cambridge. StemBook [Internet] PMID:23785737
Sanders SJ, Murtha MT, Gupta AR, Murdoch JD, Raubeson MJ, Willsey AJ, Ercan-Sencicek AG, DiLullo NM, Parikshak NN, Stein JL, Walker MF, Ober GT, Teran NA, Song Y, El-Fishawy P, Murtha RC, Choi M, Overton JD, Bjornson RD, Carriero NJ, Meyer KA, Bilguvar K, Mane SM, Sestan N, Lifton RP, Günel M, Roeder K, Geschwind DH, Devlin B, State MW (2012) De novo mutations revealed by whole-exome sequencing are strongly associated with autism. Nature 485:237–241
Sanjana NE (2016) Genome-scale CRISPR pooled screens. Anal Biochem. pii:S0003-2697(16)30089-6
Sanjana NE, Shalem O, Zhang F (2014) Improved vectors and genome-wide libraries for CRISPR screening. Nat Methods 11:783–784
Sanjana NE, Wright J, Zheng K, Shalem O, Fontanillas P, Joung J, Cheng C, Regev A, Zhang F (2016) High-resolution interrogation of functional elements in the noncoding genome. Science 353(6307):1545–1549
Shalem O, Sanjana NE, Hartenian E, Shi X, Scott DA, Mikkelsen TS, Heckl D, Ebert BL, Root DE, Doench JG, Zhang F (2014) Genome-scale CRISPR-Cas9 knockout screening in human cells. Science 343:84–87
Swiech L, Heidenreich M, Banerjee A, Habib N, Li Y, Trombetta J, Sur M, Zhang F (2015) In vivo interrogation of gene function in the mammalian brain using CRISPR-Cas9. Nat Biotechnol 33:102–106
Wright JB, Sanjana NE (2016) CRISPR screens to discover functional noncoding elements. Trends Genet 32:526–529
Zhang SC, Wernig M, Duncan ID, Brüstle O, Thomson JA (2001) In vitro differentiation of transplantable neural precursors from human embryonic stem cells. Nat Biotechnol 19:1129–1133
Zhang Y, Pak C, Han Y, Ahlenius H, Zhang Z, Chanda S, Marro S, Patzke C, Acuna C, Covy J, Xu W, Yang N, Danko T, Chen L, Wernig M, Südhof TC (2013) Rapid single-step induction of functional neurons from human pluripotent stem cells. Neuron 78:785–798
Author information
Authors and Affiliations
Corresponding author
Editor information
Editors and Affiliations
Rights and permissions
Open Access This chapter is licensed under the terms of the Creative Commons Attribution 4.0 International License (http://creativecommons.org/licenses/by/4.0/), which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license and indicate if changes were made.
The images or other third party material in this chapter are included in the chapter's Creative Commons license, unless indicated otherwise in a credit line to the material. If material is not included in the chapter's Creative Commons license and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder.
Copyright information
© 2017 The Author(s)
About this chapter
Cite this chapter
Sanjana, N.E. (2017). Multiscale Genome Engineering: Genome-Wide Screens and Targeted Approaches. In: Jaenisch, R., Zhang, F., Gage, F. (eds) Genome Editing in Neurosciences. Research and Perspectives in Neurosciences. Springer, Cham. https://doi.org/10.1007/978-3-319-60192-2_8
Download citation
DOI: https://doi.org/10.1007/978-3-319-60192-2_8
Published:
Publisher Name: Springer, Cham
Print ISBN: 978-3-319-60191-5
Online ISBN: 978-3-319-60192-2
eBook Packages: Biomedical and Life SciencesBiomedical and Life Sciences (R0)