Abstract
The repair of mammalian DNA double-strand breaks (DSBs) by classical non-homologous end joining (C-NHEJ) suppresses genomic instability and cancer and is required for development of the immune and nervous system. We hypothesize that proper repair of neural DSBs via C-NHEJ or other end-joining pathways is critical for neural functionality and homeostasis over time and that improper DSB repair could contribute to complex psychiatric and neurodegenerative diseases. Here, we summarize various findings made by our laboratory and others over the years that support this hypothesis. This evidence includes, most recently, our discovery of a set of genes, of which most serve neural functions, that can serve as targets of recurrent DSBs in primary neural stem and progenitor cells. We also present a speculative model, based on our findings, of mechanisms by which recurrent DSBs in neural genes can generate neuronal diversity and contribute to neuropsychiatric disease.
You have full access to this open access chapter, Download chapter PDF
Similar content being viewed by others
Early studies revealed that the lymphocyte-specific V(D)J recombination reaction involves the introduction of DNA double-stranded breaks (DSBs) at the ends of antigen receptor V, D, and J gene segments, followed by the processing of the generated ends and subsequent fusion of the DSB ends of the different types of gene segments to form V(D)J variable regions exons (Alt and Baltimore 1982). The Baltimore lab discovered the lymphocyte-specific endonuclease (RAG) that generates V(D)J DSBs (Schatz and Swanson 2011). Based on screens of DNA repair-mutant Chinese hamster ovary cell lines, we discovered that the end-joining phase of V(D)J recombination is carried out by a multi-component DSB end-joining pathway (Taccioli et al. 1993). We went on with collaborators to identify many of the various components of the “classical” non-homologous end-joining (C-NHEJ) pathway, including discovering the XRCC4 “core” C-NHEJ factor, based on our finding that this factor restores the ability of a DNA repair-defective Chinese hamster ovary cell line to undergo the joining phase of V(D)J recombination (Li et al. 1995).
To evaluate potential physiological functions of XRCC4 and other C-NHEJ factors newly discovered at the time, or other putative C-NHEJ factors, we inactivated the genes encoding them in mice (Sekiguchi et al. 1999; Ferguson and Alt 2001). Mice in which we inactivated the XRCC4 C-NHEJ factor, or its interaction partner DNA Ligase 4 (Lig4), had essentially identical phenotypes. These phenotypes included, most notably, abrogation of both lymphocyte and neuronal development due to unrepaired DSBs that occurred at the progenitor stage (Frank et al. 1998; Gao et al. 1998). It is striking that the development of lymphocytes and neurons was the most clear-cut defect in these C-NHEJ-deficient mice. As discussed below, XRCC4- or Lig4-deficient mice routinely die late in embryonic development, most likely due to their neuronal developmental defects. At this stage, effects on fetal lymphocyte development can still be assessed.
Lymphocyte development is blocked at the progenitor stages in these core C-NHEJ-deficient backgrounds due to the inability to join V(D)J recombination-associated DSBs generated by the RAG endonuclease in the absence of core C-NHEJ factors (Alt et al. 2013). Thus, progenitor B and T lymphocyte development was completely abrogated due to the inability to, respectively, assemble functional antibody and T cell receptor genes that are needed for further development of the B and T cell lineages. As V(D)J recombination occurs at the G1 cell cycle stage, core C-NHEJ-deficient progenitor lymphocytes correspondingly undergo apoptosis due to a response to their unrepaired V(D)J DSBs that is mediated by the p53 G1 check-point response factor (Frank et al. 2000; Gao et al. 2000; Zhu et al. 2002). In this regard, p53 deficiency, in fact, rescues the embryonic lethality of XRCC4- or Lig4-deficient mice but does not rescue lymphocyte development because V(D)J joining is still abrogated. The alleviation of the p53 response to unrepaired RAG-generated DSBs at antigen receptor genes allows XRCC4- or Lig4-deficient progenitor lymphocytes to survive and enter the cell cycle, resulting in XRCC4/p53-deficient mice that rapidly develop lethal pro-B cell lymphomas (Frank et al. 2000; Gao et al. 2000). These C-NHEJ/p53-deficient pro-B lymphomas all harbor recurrent translocations that fuse RAG-initiated DSBs at the IgH locus to DSBs downstream of c-Myc (Zhu et al. 2002), with many likely initiated at cryptic RAG off-targets sites in the c-Myc downstream region (Hu et al. 2014; Tepsuporn et al. 2014). Notably, however, even though core C-NHEJ-deficient/p53-deficient mice die from recurrent pro-B lymphomas, many of them harbor medulloblastomas in situ at the time of their death from pro-B lymphoma (Zhu et al. 2002). Finally, conditional inactivation of Xrcc4 in p53-deficient B cells leads to mature B lymphomas with recurrent translocations involving DSBs initiated by the B cell-specific activation-induced cytidine deaminase (AID) during IgH class switch recombination (CSR, see below) that are joined to upstream regions of the c-Myc gene (Wang et al. 2009).
Our studies demonstrated that XRCC4- or Lig4-deficient neuronal progenitor cells undergo apoptosis throughout the nervous system at a developmental time when particular neuronal progenitor populations differentiate into postmitotic neurons (Gao et al. 1998). Moreover, we implicated p53 checkpoint-initiated apoptosis in response to unrepaired DSBs that occurred in the neuronal progenitors as a mechanism for this death of newly differentiated neurons, as demonstrated by our finding that such neuronal apoptotic death could be rescued by p53 deficiency. In this regard, the postnatal survival of XRCC4-deficient or Lig4-deficient mice conferred by p53 deficiency has been speculated to be due to rescue of newly differentiated neurons with unrepaired DSBs (Sekiguchi et al. 1999). However, the potential effects of such unrepaired DSBs on neuronal functions in these mice could not be assessed due to their rapid death from pro-B cell lymphomas; thus, the potential roles of these implied DSBs in neuronal development and neuronal functions remained speculative. In this regard, a lingering question was the location of the genomic sites of the involved DSBs.
As mentioned above, C-NHEJ/p53 double-deficient mice all develop progenitor B cell lymphomas with recurrent translocations between the IgH and c-Myc genes, whereas p53-deficient mice with Xrcc4 conditionally inactivated in B-lineage cells develop mature B-lineage tumors with translocations between IgH and c-Myc but also translocations of other antigen receptor loci (Wang et al. 2008, 2009). Thus, we attempted to identify recurrently breaking genomic sites in neural progenitor cells by conditionally inactivating Xrcc4 in neuronal stem and progenitor cells in a p53-deficient background. Strikingly, we found that such conditional inactivation of Xrcc4 in p53-deficient neural progenitors routinely led to medulloblastomas (MBs) with recurrent translocations on several different chromosomes and frequent chromosomal or extrachromosomal amplification of the N-myc gene (Yan et al. 2006). These N-myc amplifications were reminiscent of those we found in human neuroblastomas in the process of discovering N-myc (Kohl et al. 1983). While the findings supported our original hypothesis that recurrent DSBs in the vicinity of N-myc (or other frequently translocated regions in MBs) could predispose to such translocations and amplifications, the resolution available from our studies at that time did not allow mapping of potential fragile break sites.
Together, our prior studies revealed that DSB repair by C-NHEJ in neural stem and progenitor cells (NSPCs) is required for nervous system development and for suppressing childhood brain tumors (Gao et al. 1998; Yan et al. 2006). These studies also raised the interesting possibility of potential parallels between functional outcomes of DSB generation and repair in lymphocytes and neuronal progenitor cells. More recently, studies by others have shown that mature brain cells contain frequent genomic alterations that have been speculated to contribute to neuronal diversity and disease (McConnell et al. 2013; Poduri et al. 2013; Weissman and Gage 2016). In this regard, beyond inherited germline mutations, somatic, “brain only”, mutations have been implicated in neurodevelopmental and neuropsychiatric disorders (Poduri et al. 2013). However, the potential causes of genomic alterations in brain cells continued to remain largely unexplored and speculative. Based on our observations regarding the effects of C-NHEJ deficiency on neuronal development and neuronal disease, namely cancer, we sought to develop and employ new technologies to test the hypothesis that genomic alterations in mature brain cells and some variations connected to neuropsychiatric diseases might originate from DSBs in NSPCs.
Over the past decade, since our discoveries of the potential roles for DSBs in neuronal diversity and disease, we have developed and enhanced a high-throughput, genome-wide translocation sequencing (HTGTS) approach to rapidly and highly sensitively identify DSBs genome-wide based on their translocation to bait DSBs (Chiarle et al. 2011; Frock et al. 2015; Hu et al. 2016). For this approach, bait DSBs can be introduced ectopically by designer endonucleases (Chiarle et al. 2011; Hu et al. 2014; Meng et al. 2014; Frock et al. 2015) or recurrent endogenous DSBs can be used as bait, including those initiated by AID during IgH CSR (Dong et al. 2015) or by RAG during V(D)J recombination (Zhang et al. 2012; Hu et al. 2015; Zhao et al. 2016).
Our studies have shown that various classes of DSBs, including those induced ectopically by ionizing radiation, show a much greater preference to join to other DSBs within the same topological domain due to proximity effects associated with the spatial genome organization of chromatin domains (Zarrin et al. 2007; Zhang et al. 2012; Alt et al. 2013; Frock et al. 2015). As two random DSBs rarely occur within the relatively short genomic distances within a chromosomal domain, which is often a megabase or less, this phenomenon most greatly impacts the joining of closely linked recurrent DSBs (Alt et al. 2013). Our HTGTS studies provided additional insights into our prior finding (Zarrin et al. 2007; Gostissa et al. 2014) that indicated that CSR joining exploits the predisposition of high frequency DSBs within topological domains to be joined to each other to achieve physiological joining levels (Zarrin et al. 2007; Dong et al. 2015). We also showed that, during V(D)J recombination, RAG exploits chromosomal loop domains to not only achieve high joining frequency but also to developmentally restrict its activity directionally within a loop domain (Hu et al. 2015; Zhao et al. 2016).
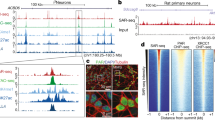
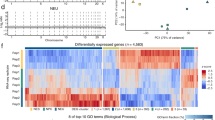
To identify the sources and functions of neural DSBs, we applied our HTGTS DSB identification approach to cultured, primary mouse NSPCs. For these HTGTS studies, we employed ectopically generated bait DSBs on several different chromosomes to search for significant, recurrent clusters of DSBs genome-wide that joined to bait DSBs on more than one chromosome. These studies identified 27 recurrent DSB clusters (“RDCs”) in the NSPC genome, all of which were enhanced by mild replication stress via treatment with aphidicolin, a compound that inhibits replication (Wei et al. 2016). Strikingly, all 27 of these RDCs lie within genes, most of which encode surface proteins involved in synaptogenesis and related neural processes (Wei et al. 2016). Moreover, variations of most RDC genes also have been implicated in neuropsychiatric disorders, including schizophrenia and autism, and many are rearranged in cancers, including brain cancers such as medulloblastoma (Wei et al. 2016; Weissman and Gage 2016). Notably, human counterparts of 9 of the 27 NSPC RDC genes occurred in copy number variations (CNVs) found in individual human frontal cortex neurons (McConnell et al. 2013), suggesting that NSPC RDC DSBs could contribute genomic variations in mature neurons (Wei et al. 2016; Weissman and Gage 2016).
RDC gene transcriptional and replication characteristics suggest that their frequent DSBs could occur during collisions between RNA and DNA polymerases associated with mild replication stress (Wei et al. 2016). RDC gene DSBs appear to occur very frequently across the body of RDC genes, which generally are very long (up to 2 Mb in length) with relatively small exons and which also potentially often lie within topological domains (Wei et al. 2016). As HTGTS maps only those bait DSBs that translocate, local RDC DSB frequency may be much higher than the estimated minimal frequency of 12 RDC translocations per NSPC that we estimated via translocation junction capture via HTGTS (Wei et al. 2016). Indeed, we have estimated that the frequency of DSBs across long RDC genes, while of lower density than CSR DSBs, approach the same order of magnitude in numbers per gene as CSR DSBs in B lymphocytes during IgH CSR (Wei et al. 2016). Notably, because most of the RDC gene sequences are within introns, most of the RDC DSBs also occur within introns as opposed to within exons (Wei et al. 2016).
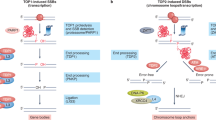
By analogy to mechanisms of lymphocyte-specific recombination (Dong et al. 2015; Hu et al. 2015), we propose that many DSBs that occur within RDC genes would be joined to other DSBs within the same RDC gene (Wei et al. 2016). Thus, we further propose that frequent RDC gene DSBs, which again mostly occur within introns, may be joined to shuffle exons and, thereby, contribute to neural cell diversity (Fig. 1). Such breakage and joining events may also have the potential of contributing to disease-associated neural gene alterations (Wei et al. 2016; Weissman and Gage 2016).
Top panel Diagram of the IgH class switch recombination reaction as illustrated by switching from IgM to IgG1. The IgH locus is contained with a topological domain (TAD). In activated B cells, switching from IgM to IgG1 results from an exon shuffling process in which the V(D)J exon is first expressed with Cμ to generate IgM but, upon activation, DSBs initiated by AID in repetitive switch (S) regions upstream of Cμ and Cγ1 are joined by C-NHEJ to delete Cμ and replace it with Cγ1. This recombination/deletion exon shuffling process allows the same V(D)J exon to be expressed with a different C exon (For other details, see text or Alt et al. 2013). Bottom Panel Diagram of a hypothetical RDC DSB-based exon shuffling mechanism to allow expression of different isoforms of RDC genes to be expressed by “hardwiring” potential somatic splice variants by deletional recombination. This model is based on the finding that at least some RDC genes lie within TADs and that RDC DSB frequency upon replication stress may approach that of IgH S regions, allowing ends of different RDC DSBs within the same gene to be frequently joined, based on their proximity within the same topological domain. This model could offer one explanation for why many neural genes are very large and embedded with relatively small exons (Smith et al. 2006): namely, as these genes are mostly comprised of intronic sequences, most “randomly” introduced RDC DSBs across them fall within intronic sequences rather than in exons, providing a basis for a replication stress-associated DSB diversification mechanism. If so, whether or not requisite replication stress is somehow programmed during NSPC development remains to be addressed (See text or Wei et al. 2016 for other details)
A number of RDC genes, for example, the neurexins (Treutlein et al. 2014), are thought to produce numerous isoforms via differential RNA processing. Beyond such a diversification mechanism, we propose that RDC-based recombination, by generating exon deletions, might “hard-wire” expression of variant RDC products in NSPCs and, thereby, contribute to neural diversity. Our current findings suggest that such putative activities would occur in NSPCs and the products of recombination events would be carried on into mature neurons; in this regard, the process would be somewhat analogous to V(D)J recombination. However, the actual exon shuffling mechanism we propose would be more similar to IgH CSR, creating different isoforms of the protein rather than creating new exons (Fig. 1). In this scenario, the evolution of long, neural genes that are largely comprised of intronic sequences into which are embedded small exons (Smith et al. 2006) could have evolved to provide large target introns for more random stress-associated DSBs in NSPC development. This would be a different solution to the problem of targeted exon shuffling than that employed by CSR, in which DSBs are introduced into specialized intronic switch region sequences (Fig. 1). Whether or not the processes that generate RDC genes are specialized to the neural lineage will require further investigation, as will the question of whether enhanced replication stress at the stem and progenitor development stages during neural development could, via an RDC-based mechanism, contribute to neural disease.
RDCs also potentially provide a mechanistic basis for many common fragile sites and certain CNVs, which may result from transcription/replication collisions in generating DSBs or other lesions (Glover and Wilson 2016; Wei et al. 2016). Two NSPC-RDC genes, CDH13 and NRXN3, are within recurrent CNVs in human MBs (Northcott et al. 2012; Rausch et al. 2012) and several candidate RDCs lie proximal to mouse N-myc (Wei, Schwer and Alt, unpublished data). It is possible that RDCs contribute to recurrent genomic variations we and others have found in MBs (Yan et al. 2006), which may offer a mechanism to support the speculation from long ago that proximal, recurrent DSBs during neuroblast differentiation contribute to N-myc amplification in human neuroblastomas (Kohl et al. 1983). A number of the 27 identified NSPC RDC-genes undergo somatic genomic rearrangements, including deletions, amplifications, and translocations in various types of cancer (see Wei et al. 2016), and some undergo CNVs in embryonic stem cells and fibroblasts (Wilson et al. 2015; Glover and Wilson 2016). Our HTGTS analysis of additional cell types could identify potential spontaneous or replication stress-induced RDCs in other cell types and, more generally, could shed light on the mechanisms underlying the genetic variations in a range of cancers.
References
Alt FW, Baltimore D (1982) Joining of immunoglobulin heavy chain gene segments: implications from a chromosome with evidence of three D-JH fusions. Proc Natl Acad Sci USA 79:4118–4122
Alt FW, Zhang Y, Meng FL, Guo C, Schwer B (2013) Mechanisms of programmed DNA lesions and genomic instability in the immune system. Cell 152:417–429
Chiarle R, Zhang Y, Frock RL, Lewis SM, Molinie B, Ho YJ, Myers DR, Choi VW, Compagno M, Malkin DJ, Neuberg D, Monti S, Giallourakis CC, Gostissa M, Alt FW (2011) Genome-wide translocation sequencing reveals mechanisms of chromosome breaks and rearrangements in B cells. Cell 147:107–119
Dong J, Panchakshari RA, Zhang T, Zhang Y, Hu J, Volpi SA, Meyers RM, Ho YJ, Du Z, Robbiani DF, Meng F, Gostissa M, Nussenzweig MC, Manis JP, Alt FW (2015) Orientation-specific joining of AID-initiated DNA breaks promotes antibody class switching. Nature 525:134–139
Ferguson DO, Alt FW (2001) DNA double strand break repair and chromosomal translocation: lessons from animal models. Oncogene 20:5572–5579
Frank KM, Sekiguchi JM, Seidl KJ, Swat W, Rathbun GA, Cheng HL, Davidson L, Kangaloo L, Alt FW (1998) Late embryonic lethality and impaired V(D)J recombination in mice lacking DNA ligase IV. Nature 396:173–177
Frank KM, Sharpless NE, Gao Y, Sekiguchi JM, Ferguson DO, Zhu C, Manis JP, Horner J, DePinho RA, Alt FW (2000) DNA ligase IV deficiency in mice leads to defective neurogenesis and embryonic lethality via the p53 pathway. Mol Cell 5:993–1002
Frock RL, Hu J, Meyers RM, Ho YJ, Kii E, Alt FW (2015) Genome-wide detection of DNA double-stranded breaks induced by engineered nucleases. Nat Biotechnol 33:179–186
Gao Y, Sun Y, Frank KM, Dikkes P, Fujiwara Y, Seidl KJ, Sekiguchi JM, Rathbun GA, Swat W, Wang J, Bronson RT, Malynn BA, Bryans M, Zhu C, Chaudhuri J, Davidson L, Ferrini R, Stamato T, Orkin SH, Greenberg ME, Alt FW (1998) A critical role for DNA end-joining proteins in both lymphogenesis and neurogenesis. Cell 95:891–902
Gao Y, Ferguson DO, Xie W, Manis JP, Sekiguchi J, Frank KM, Chaudhuri J, Horner J, DePinho RA, Alt FW (2000) Interplay of p53 and DNA-repair protein XRCC4 in tumorigenesis, genomic stability and development. Nature 404:897–900
Glover TW, Wilson TE (2016) Molecular biology: breaks in the brain. Nature 532:46–47
Gostissa M, Schwer B, Chang A, Dong J, Meyers RM, Marecki GT, Choi VW, Chiarle R, Zarrin AA, Alt FW (2014) IgH class switching exploits a general property of two DNA breaks to be joined in cis over long chromosomal distances. Proc Natl Acad Sci USA 111:2644–2649
Hu J, Tepsuporn S, Meyers RM, Gostissa M, Alt FW (2014) Developmental propagation of V(D)J recombination-associated DNA breaks and translocations in mature B cells via dicentric chromosomes. Proc Natl Acad Sci USA 111:10269–10274
Hu J, Zhang Y, Zhao L, Frock RL, Du Z, Meyers RM, Meng FL, Schatz DG, Alt FW (2015) Chromosomal loop domains direct the recombination of antigen receptor genes. Cell 163:947–959
Hu J, Meyers RM, Dong J, Panchakshari RA, Alt FW, Frock RL (2016) Detecting DNA double-stranded breaks in mammalian genomes by linear amplification-mediated high-throughput genome-wide translocation sequencing. Nat Protoc 11:853–871
Kohl NE, Kanda N, Schreck RR, Bruns G, Latt SA, Gilbert F, Alt FW (1983) Transposition and amplification of oncogene-related sequences in human neuroblastomas. Cell 35:359–367
Li Z, Otevrel T, Gao Y, Cheng HL, Seed B, Stamato TD, Taccioli GE, Alt FW (1995) The XRCC4 gene encodes a novel protein involved in DNA double-strand break repair and V(D)J recombination. Cell 83:1079–1089
McConnell MJ, Lindberg MR, Brennand KJ, Piper JC, Voet T, Cowing-Zitron C, Shumilina S, Lasken RS, Vermeesch JR, Hall IM, Gage FH (2013) Mosaic copy number variation in human neurons. Science 342:632–637
Meng FL, Du Z, Federation A, Hu J, Wang Q, Kieffer-Kwon KR, Meyers RM, Amor C, Wasserman CR, Neuberg D, Casellas R, Nussenzweig MC, Bradner JE, Liu XS, Alt FW (2014) Convergent transcription at intragenic super-enhancers targets AID-initiated genomic instability. Cell 159:1538–1548
Northcott PA, Shih DJ, Peacock J, Garzia L, Morrissy AS, Zichner T, Stütz AM, Korshunov A, Reimand J, Schumacher SE, Beroukhim R, Ellison DW, Marshall CR, Lionel AC, Mack S, Dubuc A, Yao Y, Ramaswamy V, Luu B, Rolider A, Cavalli FM, Wang X, Remke M, Wu X, Chiu RY, Chu A, Chuah E, Corbett RD, Hoad GR, Jackman SD, Li Y, Lo A, Mungall KL, Nip KM, Qian JQ, Raymond AG, Thiessen NT, Varhol RJ, Birol I, Moore RA, Mungall AJ, Holt R, Kawauchi D, Roussel MF, Kool M, Jones DT, Witt H, Fernandez-L A, Kenney AM, Wechsler-Reya RJ, Dirks P, Aviv T, Grajkowska WA, Perek-Polnik M, Haberler CC, Delattre O, Reynaud SS, Doz FF, Pernet-Fattet SS, Cho BK, Kim SK, Wang KC, Scheurlen W, Eberhart CG, Fèvre-Montange M, Jouvet A, Pollack IF, Fan X, Muraszko KM, Gillespie GY, Di Rocco C, Massimi L, Michiels EM, Kloosterhof NK, French PJ, Kros JM, Olson JM, Ellenbogen RG, Zitterbart K, Kren L, Thompson RC, Cooper MK, Lach B, McLendon RE, Bigner DD, Fontebasso A, Albrecht S, Jabado N, Lindsey JC, Bailey S, Gupta N, Weiss WA, Bognár L, Klekner A, Van Meter TE, Kumabe T, Tominaga T, Elbabaa SK, Leonard JR, Rubin JB, Liau LM, Van Meir EG, Fouladi M, Nakamura H, Cinalli G, Garami M, Hauser P, Saad AG, Iolascon A, Jung S, Carlotti CG, Vibhakar R, Ra YS, Robinson S, Zollo M, Faria CC, Chan JA, Levy ML, Sorensen PH, Meyerson M, Pomeroy SL, Cho YJ, Bader GD, Tabori U, Hawkins CE, Bouffet E, Scherer SW, Rutka JT, Malkin D, Clifford SC, Jones SJ, Korbel JO, Pfister SM, Marra MA, Taylor MD (2012) Subgroupspecific structural variation across 1,000 medulloblastoma genomes. Nature 488:49–56
Poduri A, Evrony GD, Cai X, Walsh CA (2013) Somatic mutation, genomic variation, and neurological disease. Science 341:1237758
Rausch T, Shih DJ, Peacock J, Garzia L, Morrissy AS, Zichner T, Stütz AM, Korshunov A, Reimand J, Schumacher SE, Beroukhim R, Ellison DW, Marshall CR, Lionel AC, Mack S, Dubuc A, Yao Y, Ramaswamy V, Luu B, Rolider A, Cavalli FM, Wang X, Remke M, Wu X, Chiu RY, Chu A, Chuah E, Corbett RD, Hoad GR, Jackman SD, Li Y, Lo A, Mungall KL, Nip KM, Qian JQ, Raymond AG, Thiessen NT, Varhol RJ, Birol I, Moore RA, Mungall AJ, Holt R, Kawauchi D, Roussel MF, Kool M, Jones DT, Witt H, Fernandez-L A, Kenney AM, Wechsler-Reya RJ, Dirks P, Aviv T, Grajkowska WA, Perek-Polnik M, Haberler CC, Delattre O, Reynaud SS, Doz FF, Pernet-Fattet SS, Cho BK, Kim SK, Wang KC, Scheurlen W, Eberhart CG, Fèvre-Montange M, Jouvet A, Pollack IF, Fan X, Muraszko KM, Gillespie GY, Di Rocco C, Massimi L, Michiels EM, Kloosterhof NK, French PJ, Kros JM, Olson JM, Ellenbogen RG, Zitterbart K, Kren L, Thompson RC, Cooper MK, Lach B, McLendon RE, Bigner DD, Fontebasso A, Albrecht S, Jabado N, Lindsey JC, Bailey S, Gupta N, Weiss WA, Bognár L, Klekner A, Van Meter TE, Kumabe T, Tominaga T, Elbabaa SK, Leonard JR, Rubin JB, Liau LM, Van Meir EG, Fouladi M, Nakamura H, Cinalli G, Garami M, Hauser P, Saad AG, Iolascon A, Jung S, Carlotti CG, Vibhakar R, Ra YS, Robinson S, Zollo M, Faria CC, Chan JA, Levy ML, Sorensen PH, Meyerson M, Pomeroy SL, Cho YJ, Bader GD, Tabori U, Hawkins CE, Bouffet E, Scherer SW, Rutka JT, Malkin D, Clifford SC, Jones SJ, Korbel JO, Pfister SM, Marra MA, Taylor MD (2012) Genome sequencing of pediatric medulloblastoma links catastrophic DNA rearrangements with TP53 mutations. Cell 148:59–71
Schatz DG, Swanson PC (2011) V(D)J recombination: mechanisms of initiation. Annu rev Genet 45:167–202
Sekiguchi JM, Gao Y, Gu Y, Frank K, Sun Y, Chaudhuri J, Zhu C, Cheng HL, Manis J, Ferguson D, Davidson L, Greenberg ME, Alt FW (1999) Nonhomologous end-joining proteins are required for V(D)J recombination, normal growth, and neurogenesis. Cold Spring Harb Symp Quant Biol 64:169–181
Smith DI, Zhu Y, McAvoy S, Kuhn R (2006) Common fragile sites, extremely large genes, neural development and cancer. Cancer Lett 232:48–57
Taccioli GE, Rathbun G, Oltz E, Stamato T, Jeggo PA, Alt FW (1993) Impairment of V(D)J recombination in double-strand break repair mutants. Science 260:207–210
Tepsuporn S, Hu J, Gostissa M, Alt FW (2014) Mechanisms that can promote peripheral B-cell lymphoma in ATM-deficient mice. Cancer Immunol res 2:857–866
Treutlein B, Gokce O, Quake SR, Sudhof TC (2014) Cartography of neurexin alternative splicing mapped by single-molecule long-read mRNA sequencing. Proc Natl Acad Sci USA 111:E1291–E1299
Wang JH, Alt FW, Gostissa M, Datta A, Murphy M, Alimzhanov MB, Coakley KM, Rajewsky K, Manis JP, Yan CT (2008) Oncogenic transformation in the absence of Xrcc4 targets peripheral B cells that have undergone editing and switching. J Exp med 205:3079–3090
Wang JH, Gostissa M, Yan CT, Goff P, Hickernell T, Hansen E, Difilippantonio S, Wesemann DR, Zarrin AA, Rajewsky K, Nussenzweig A, Alt FW (2009) Mechanisms promoting translocations in editing and switching peripheral B cells. Nature 460:231–236
Wei PC, Chang AN, Kao J, Du Z, Meyers RM, Alt FW, Schwer B (2016) Long neural genes harbor recurrent DNA break clusters in neural stem/progenitor cells. Cell 164:644–655
Weissman IL, Gage FH (2016) A mechanism for somatic brain mosaicism. Cell 164:593–595
Wilson TE, Arlt MF, Park SH, Rajendran S, Paulsen M, Ljungman M, Glover TW (2015) Large transcription units unify copy number variants and common fragile sites arising under replication stress. Genome res 25:189–200
Yan CT, Kaushal D, Murphy M, Zhang Y, Datta A, Chen C, Monroe B, Mostoslavsky G, Coakley K, Gao Y, Mills KD, Fazeli AP, Tepsuporn S, Hall G, Mulligan R, Fox E, Bronson R, De Girolami U, Lee C, Alt FW (2006) XRCC4 suppresses medulloblastomas with recurrent translocations in p53-deficient mice. Proc Natl Acad Sci USA 103:7378–7383
Zarrin AA, Del Vecchio C, Tseng E, Gleason M, Zarin P, Tian M, Alt FW (2007) Antibody class switching mediated by yeast endonuclease-generated DNA breaks. Science 315:377–381
Zhang Y, McCord RP, Ho YJ, Lajoie BR, Hildebrand DG, Simon AC, Becker MS, Alt FW, Dekker J (2012) Spatial organization of the mouse genome and its role in recurrent chromosomal translocations. Cell 148:908–921
Zhao L, Frock RL, Du Z, Hu J, Chen L, Krangel MS, Alt FW (2016) Orientation-specific RAG activity in chromosomal loop domains contributes to Tcrd V(D)J recombination during T cell development. J Exp med 213:1921–1936
Zhu C, Mills KD, Ferguson DO, Lee C, Manis J, Fleming J, Gao Y, Morton CC, Alt FW (2002) Unrepaired DNA breaks in p53-deficient cells lead to oncogenic gene amplification subsequent to translocations. Cell 109:811–821
Acknowledgments
This work was supported by the Porter Anderson Fund from Boston Children’s Hospital and the Howard Hughes Medical Institute. P.W. was supported by a National Cancer Center postdoctoral fellowship. B.S. is a Martin D. Abeloff Scholar of The V Foundation for Cancer Research and is supported by NIA/NIH grant K01AG043630.
Author information
Authors and Affiliations
Corresponding author
Editor information
Editors and Affiliations
Rights and permissions
Open Access This chapter is licensed under the terms of the Creative Commons Attribution 4.0 International License (http://creativecommons.org/licenses/by/4.0/), which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license and indicate if changes were made.
The images or other third party material in this chapter are included in the chapter's Creative Commons license, unless indicated otherwise in a credit line to the material. If material is not included in the chapter's Creative Commons license and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder.
Copyright information
© 2017 The Author(s)
About this chapter
Cite this chapter
Alt, F.W., Wei, PC., Schwer, B. (2017). Recurrently Breaking Genes in Neural Progenitors: Potential Roles of DNA Breaks in Neuronal Function, Degeneration and Cancer. In: Jaenisch, R., Zhang, F., Gage, F. (eds) Genome Editing in Neurosciences. Research and Perspectives in Neurosciences. Springer, Cham. https://doi.org/10.1007/978-3-319-60192-2_6
Download citation
DOI: https://doi.org/10.1007/978-3-319-60192-2_6
Published:
Publisher Name: Springer, Cham
Print ISBN: 978-3-319-60191-5
Online ISBN: 978-3-319-60192-2
eBook Packages: Biomedical and Life SciencesBiomedical and Life Sciences (R0)