Abstract
This review updates the previous information about ectomycorrhizal (EcM) fungal lineages. Based on morphological and phylogenetic evidence, we recognize four novel lineages, viz., /leotia, /phaeocollybia, /porpoloma, and /endogone3, that raise the total number of EcM lineages to 82–86. The /leotia, /phaeocollybia, and /porpoloma lineages comprise the entire genera of Leotia, Phaeocollybia, and Porpoloma. The /endogone3 lineage is created to accommodate morphologically, and molecularly well-characterized root tip isolates from Japan that represent a small proportion of the genus Endogone. The /agaricales1 lineage is revised and renamed /guyanagarika because of accommodation of respective specimens in a recently described genus Guyanagarika. Similarly, the /helotiales2 lineage is renamed as /phaeohelotium based description of the genus Phaeohelotium. In light of broader taxon sampling for fungal DNA barcoding, the /agaricomycetes1 lineage is renamed as /tremellodendropsis, with a notion that not all members of the Tremellodendropsidales are EcM. We also reviewed recent literature about the genera within described EcM lineages and documented that ca. 279–284 distinct EcM fungal genera based on current knowledge.
Because EcM symbionts are now frequently identified from soil and roots using high-throughput sequencing methods, we also provide recommendations for delimitation of each lineage in relation to non-mycorrhizal sister taxa by integrating information from BLASTn score, sequence length, and sequence similarity. These models can be used as a guide to assign sequences to EcM versus non-EcM taxa for most lineages, but some individual lineages require additional sequence similarity and/or phylogenetic analyses for reliable placement due to rapid ITS evolution. We conclude that efficient automatized identification and functional assignment of fungal taxa requires integration of taxonomic, functional, and genomic information at the level of Linnaean taxa as well as species hypotheses.
Similar content being viewed by others
Keywords
- Ectomycorrhizal symbiosis
- Molecular identification
- Next-generation sequencing
- Metabarcoding
- Taxonomic identification
- Taxonomic assignment algorithms
- Sequence metadata
- UNITE database
- Leotia
- Porpoloma
- Phaeocollybia
- Endogonales
6.1 Introduction
Ectomycorrhizal (EcM) fungi represent a diverse group that forms mutualistic associations with plant roots. Due to different opinions and methods, there has been significant controversy in “separating the wheat from the chaff” when assigning mycorrhizal status to fungal species or Operational Taxonomic Units (OTUs) that are recovered from molecular identification studies (Rinaldi et al. 2008; Tedersoo et al. 2010; Tedersoo and Smith 2013). Based on phylogenetic information, the EcM fungal species and genera have been grouped into monophyletic “lineages” to reflect their independent evolution from non-mycorrhizal ancestors (Tedersoo et al. 2010). Accumulating evidence suggests that this is a unidirectional process by which mostly saprobic ancestors transition into a symbiotic lifestyle. These ectomycorrhizal biotrophic fungi subsequently lose the genes responsible for plant cell wall degradation (e.g., Kohler et al. 2015), and thus reversals to saprotrophy or other trophic lifestyles are rare, nonexistent, or transient.
Using sequence metadata as well as phylogenetic and statistical analyses, Tedersoo and Smith (2013) added additional lineages of previously unrecognized EcM fungi that were only known from sequence data obtained from plant roots and/or soil. These reports increased the number of EcM lineages to 78–82. Since 2013, a number of revealing molecular identification and phylogenetic studies have been published that motivated us to revise the EcM fungal lineages in order to match the most recent knowledge.
Molecular identification studies of fungi from soil typically rely on the best BLASTn matches or Naïve Bayesian Classifier (Porras-Alfaro et al. 2014) to assign representative sequences of OTUs to species, genera, families, and higher taxonomic ranks based on subjective similarity thresholds (Tedersoo and Nilsson 2016). Both traditional Sanger sequencing and high-throughput sequencing (HTS) studies usually fail to account for the fact that the ITS regions (as well as other molecular markers) differ in their rate of evolution and therefore in the level of separation between species and across lineages. For example, it is likely that an OTU with 95% full-length ITS sequence match to the taxon Russula vinosa represents an ectomycorrhizal species in the genus Russula, but the same is not necessarily true for Cenococcum geophilum or Meliniomyces bicolor. What can we conclude about the trophic mode of OTUs from soil or roots that match R. vinosa or any other EcM fungal taxon at 80%, 85%, or 90% similarity? Inclusion or exclusion of these taxa may strongly bias the view of the EcM to saprotroph ratio and the environmental effects on fungal guilds if these OTUs are highly abundant.
Although macroscopic EcM fungi are relatively well studied compared to some other fungal groups, molecular ecology studies in tropical ecosystems or in the Southern Hemisphere commonly encounter problems in identification due to a dearth of well-annotated reference sequences from identified specimens, axenic cultures, or EcM roots. If the studies are to compare overall fungal diversity, this is not a significant problem. However, trophic groups of fungi respond to different predictors and display different biogeographic patterns. Therefore, most studies attempt to separate EcM fungi, arbuscular mycorrhizal (AM) fungi, and putative plant pathogens from potential saprotrophs. So far, the assignment of a trophic status has been typically performed based on taxonomic assignments either manually or in a semiautomatic fashion (Nguyen et al. 2016). Although ecological traits should be clearly related to collections or at least reference OTUs or species hypotheses (SHs; Kõljalg et al. 2016) rather than genus or family names, there is currently no annotated system for rigorously incorporating additional information on important ecological, morphological, and physiological traits (e.g., EcM exploration type, fruit body type, enzymatic capacities, etc.). This means that most data sets require time-consuming manual trophic assignments based on expert knowledge in order to extract critical ecological details. The current system also renders the correctness of taxonomic labeling and specimen identification of great analytical importance. Hence, we aim to assign information about EcM fungal lineages to individual isolates (accessions) and SHs in UNITE and to establish group-specific ITS sequence similarity thresholds for delimiting EcM fungal lineages based on our previous experience with high-throughput sequencing.
6.2 Approaches
We critically evaluated recent studies about the phylogeny and molecular identification of EcM fungi published since 2013. We also rechecked the sequences and metadata accumulated in the International Nucleotide Sequence Databases consortium (INSDc) and UNITE over the same time period. Lastly, we ran simple maximum likelihood phylogenetic analyses as described in Tedersoo and Smith (2013) to establish the monophyly of putative EcM groups.
To reproducibly separate EcM fungi from non-mycorrhizal fungi in HTS studies, we compiled information about the BLASTn identification of soil fungi based on the ITS2 subregion in the 454 pyrosequencing (Tedersoo et al. 2014a, 2016a) and Illumina MiSeq (Tedersoo et al. 2015a, b) HTS data sets. We also added unpublished data targeting the full ITS region that was obtained by combining primers ITS9MUNngs and ITS4ngsUni (Tedersoo and Lindahl 2016) and Pacific Biosciences RS II platform for a subset of soil samples collected from Estonia and Australia. This approach is built on the inherent assumption that EcM fungi are monophyletic groups that are separated from non-mycorrhizal relatives and that EcM lineages display a “phylogenetic gap” compared with non-mycorrhizal sister taxa. There is ample evidence for this phenomenon in phylogenetic studies, where EcM lineages are usually separated from other non-EcM taxa with relatively strong statistical support and great phylogenetic distances (e.g., a long stem). The first author has used this approach in multiple studies published since 2014. Elaborating on this further and releasing this information was motivated by the urge to make interpretation of high-throughput sequencing data more reliable.
To be able to recognize these phylogenetic gaps and separate EcM groups from non-EcM taxa, we used both accumulated ITS Sanger sequence data and HTS data. Briefly, we compiled publicly available Sanger sequences from all EcM lineages and their putative sister groups (Tedersoo et al. 2011a; Tedersoo and Smith 2013) as references. Using the above HTS data sets, we established multiple statistical indices based on sequence length, sequence coverage, and BLASTn score. We studied the distribution of these metrics in different lineages and also in certain related groups that matched best to particular EcM lineages. Among multiple candidates, we selected “BLASTn score to query sequence length ratio (S/L ratio)” and “sequence identity (%)” as the most promising indices that display the most pronounced gap between EcM and non-EcM groups. We verified the results using additional BLASTn searches, retrieving the 100 best matches and/or via phylogenetic analyses (cf. Tedersoo and Smith 2013).
6.3 Additional EcM Fungal Lineages
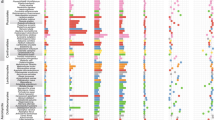
The /leotia lineage is erected to accommodate the genus Leotia of Helotiales (Fig. 6.1). Kühdorf et al. (2015) demonstrated that certain species in the genus Leotia are common root symbionts that form arbutoid EcM of short-distance exploration type with Comarostaphylis in Costa Rica. Their description of a plectenchymatous mantle of narrow clampless hyphae and thick-walled emanating hyphae roughly matches the descriptions of EcM of various groups of Helotiales. The authors also showed that a disproportionate amount of environmental sequences affiliated with the EcM group originate from Sanger-sequenced ectomycorrhizal root tips. In our previous studies, we had not noticed this sequence grouping in Leotia and thus considered this group as non-EcM based on unconfirmed root tip data from Zambia and Australia (cf. Tedersoo et al. 2010). However, earlier assessments based on isotopic evidence had previously provided suggestive evidence for an EcM habit in Leotia (Zeller et al. 2007). So far, EcM associations have only been convincingly shown for a single clade that comprises the L. lubrica and L. viscosa species complexes (Kühdorf et al. 2015). We ran a maximum likelihood phylogeny by including all Sanger sequences from fruit bodies, EcM root tips, and soil affiliated to Leotia spp. and demonstrate that most species of Leotia are likely ectomycorrhizal (Fig. 6.1). This analysis indicates that the /leotia lineage is widely distributed in all continents except perhaps lowland South America. Leotia spp. associate with Pinaceae, Fagales, Arbutoideae, Uapaca, and putatively with the Berlinia group (Fabales) and Dipterocarpaceae in Africa and India (Tedersoo et al. 2014a).
Phylogenetic placement of sequences from ectomycorrhizal root tips (EcM; in bold) and soil (data from Tedersoo et al. 2014a highlighted) among Leotia species as based on fruit bodies. Bar, 0.05 changes per position. The ITS phylogram consists of 80 terminal taxa and 603 aligned positions, with Thuemenidium atropurpureum and Microglossum viride representing an outgroup based on closest BLASTn matches to Leotia spp. Note the unexpectedly high taxonomic diversity in the earliest diverging African clade
The /porpoloma lineage is separated from /tricholoma based on the results of a multigene phylogenetic analysis by Sánchez-García et al. (2014). The /porpoloma lineage is comprised of a core group of Porpoloma, but excludes several species that have been transferred to other segregate genera: P. umbrosum and P. metapodium to Pseudotricholoma, P. spinulosum and P. macrocephalum to Pogonoloma, P. bambusarium to Corneriella, and P. pes-caprae to Pseudoporpoloma (Sánchez-García et al. 2014; Vizzini et al. 2016). As currently circumscribed, /porpoloma is a Southern Hemisphere lineage that is found in southern South America, Australia, and New Zealand. The root tip sequences originally assigned to /tricholoma (UDB002748 from Pomaderris apetala: Tedersoo et al. 2008; UDB007061 from Nothofagus dombeyi and UDB007096 and UDB007123 from N. obliqua: Nouhra et al. 2013) actually represent /porpoloma. Sequences belonging to /porpoloma have been also recovered from Nothofagus nervosa seedlings by Fernández et al. (2013) in Argentina (KJ701291). Microscopic studies of the Tasmanian and Patagonian material suggest that EcM of Porpoloma are similar to that of /tricholoma with a plectanchymatous mantle and hairy rhizomorphs that place it to the medium-distance fringe exploration type. Pseudoporpoloma pes-caprae represents a European grassland-inhabiting species that forms a sister group to the genus Tricholoma of the /tricholoma lineage (Sánchez-García et al. 2014; Vizzini et al. 2016). Following separation of Porpoloma, we treat the /tricholoma lineage as consisting only of Tricholoma species. The /tricholoma lineage is distributed in both the Northern and Southern hemispheres as well as tropical mountain regions with Fagales and Pinales and putatively with Dicymbe in the Guiana Shield region (M. E. Smith, personal observation).
The /phaeocollybia lineage is erected to accommodate species of Phaeocollybia. Three species of Phaeocollybia were reported from the roots of Abies religiosa in Mexico (Argüelles-Moyao et al. 2017), although the morphology of the ectomycorrhizas was not described. Many previous studies performed in the habitats of Phaeocollybia in North America have not detected this group on root tips. Although stable isotopes suggested the potential EcM or other biotrophic habit for Phaeocollybia spp., we previously considered this group non-mycorrhizal, because of long pseudorhizas being attached to long roots deep in soil (Redhead and Malloch 1985). The highest species diversity of Phaeocollybia (approximately 90 spp.) occurs in the temperate rainforests of western North America (Pacific Northwest), but individual endemic species are known from many regions, including Turkey, China, and northern South America (Brazil, Columbia; Coimbra et al. 2012). More work is needed to confirm that Phaeocollybia is a monophyletic group, to ensure that all species form EcM, and to document the morphology and exploration types of the symbiotic association.
We propose a novel lineage /endogone3 within Mucoromycotina based on molecular identification of Endogone sp. (accessions LC159474-LC159479) from Quercus spp. root tips and anatomical descriptions of the association (Yamamoto et al. 2017). These samples comprise a novel lineage because they represent a sister group to the saprotrophic Endogone pisiformis (Berch and Fortin 1983a, b; Berch and Castellano 1986). They are also distantly related to the /endogone1 lineage (represented by E. flammicorona and E. lactiflua), /endogone2 (E. aggregata, E. tuberculosa, Sclerogone eucalypti) and /densospora (Densospora spp.) in a multigene phylogeny of Yamamoto et al. (2017). Unfortunately, the ITS sequences were not produced, which renders DNA barcoding-based identification of this group problematic. There are also no fruit bodies matching the sequences of these collections, and, therefore, the taxonomic identity and distribution of the /endogone3 lineage remain unknown.
6.4 New Names for Previously Known EcM Lineages
The /guyanagarika lineage is created here to accommodate the lineage previously referred to by Tedersoo and Smith (2013) as /agaricales1. The genus Guyanagarika was recently erected by Sánchez-García et al. (2016) and includes only three closely related species that all occur in the Guiana Shield region of northern South America. No sequences or sporocarps from these taxa have been collected or detected outside of this region, suggesting that this may be a narrowly endemic lineage that has evolved in the Neotropics and is restricted to endemic EcM host trees such as species of Dicymbe and Pakaraimaea. The robust multi-locus phylogenetic analysis by Sánchez-García et al. (2016) placed this lineage within an expanded Catathelasmataceae but clearly separated from the members of the /catathelasma EcM lineage.
The /phaeohelotium lineage is erected to accommodate the /helotiales2 lineage that is naturally found only in the Southern Hemisphere. Dr. P. Johnston (unpubl.) first released sequences from fruit bodies of Discinella terrestris in New Zealand that matched closely to sequences from EcM root tips in Tasmania. The type species D. boudieri is only distantly related to the D. terrestris species complex, so D. terrestris was transferred to the new genus Phaeohelotium (Baral et al. 2013). The four described Phaeohelotium species are known from New Zealand and Australia and have also been documented in eucalypt plantations in Spain. Baral et al. (2013) also pointed to the observations of Warcup (1990a) that fruit bodies of D. terrestris sensu lato commonly co-occurred with other pyrophilic EcM and saprotrophic fungi after wildfire in Australia.
The /tremellodendropsis lineage is generated to accommodate the previously described /agaricomycetes1 lineage. This EcM lineage was initially erected to cover a cohesive group of Basidiomycota detected from EcM root tips especially from the Southern Hemisphere (Tedersoo and Smith 2013). A very recent fungal DNA barcoding initiative enabled to match these sequences to undescribed species of Tremellodendropsis from the formally monotypic order Tremellodendropsidales (Truong et al. 2017). This order forms a successive sister to Phallomycetidae, Stereopsidales, and Clavulicium macounii (Berbee et al. 2016). As discussed in Tedersoo and Smith (2013), not all putative species of Tremellodendropsidales are ectomycorrhizal.
6.5 Recently Revised Ectomycorrhizal Fungal Lineages
The /cenococcum lineage was discussed by Tedersoo et al. (2010) and Tedersoo and Smith (2013) as likely a group with only a few species and for which the sister taxon was poorly resolved. However, Spatafora et al. (2012) resolved several major lineages within /cenococcum and identified this lineage as belonging to Gloniaceae (with species of Glonium as the closest relatives). More recently, Obase et al. (2016) described the non-EcM Pseudocenococcum floridanum as a sister taxon to Cenococcum (see also Chap. 14).
In Mucoromycota, Tedersoo and Smith (2013) considered three EcM fungal lineages, viz., /endogone1, /endogone2, and /densospora. Because the two latter lineages had no fruit body sequences available, there was no information about their true taxonomic affinities. Our sequencing of Australian-type material (Tedersoo et al. 2016b) and recent phylogenetic analysis by Yamamoto et al. (2015) revealed that the EcM root tip sequences putatively assigned to the /endogone2 lineage are actually affiliated with Densospora in the /densospora lineage. Probably not all species of the genus Densospora form EcM (Warcup 1985; McGee 1996). The genus Sphaerocreas is also closely related to Densospora and affiliated EcM sequences, but its ecology is not well understood (Hirose et al. 2014). The /endogone2 lineage is comprised of the Australian species Endogone tuberculosa, E. aggregata, and potentially Sclerogone eucalypti (Tedersoo and Smith 2013). Specimens of EcM species Endogone aggregata and E. magnospora nom. nud. (a putative member of this group) were recently sequenced, but these do not match closely to any sequences from EcM root tips. Specimens of E. tuberculosa and S. eucalypti have not yet been sequenced due to the age and paucity of herbarium materials. Furthermore, fruit body specimens and root tips of Endogone and Densospora are problematic to amplify and sequence because of multiple divergent ITS copies and long homopolymers (Tedersoo et al. 2016b). Endogonales resemble Glomeromycota (recently proposed as Glomeromycotina within Mucoromycota; Spatafora et al. 2016) in that they form nonseptate, multinucleate hyphae. This has been best demonstrated in pure cultures of E. pisiformis (Jabaji-Hare and Charest 1987). In the /endogone1 lineage, only members of the E. flammicorona and E. lactiflua species complexes (Endogone group B sensu Yamamoto et al. 2015) have been shown to form EcM (Warcup 1990b). Unfortunately, direct molecular evidence of EcM colonization by species in the /endogone1 and /endogone2 lineages is still lacking. The trophic status and ecophysiology of Endogonales requires urgent attention, because multiple distant clades of this group are likely recognized in the morphological species “Glomus tenue” s. lat. These have been referred to as “fine endophytes” that routinely colonize roots and form arbuscule-like structures in AM vascular plants (Orchard et al. 2017) and coils of hyphae in liverworts (Field et al. 2015).
6.6 Potential EcM Lineages that Require More Data
Several EcM lineages or putative EcM lineages still require more sampling effort to elucidate their interactions with host plants or clarify their putative trophic modes. For some of these taxa, their EcM status is suggested by the fruiting habit, associations with host plants, and/or isotopic data. However, for several groups we still lack solid data on EcM morphology and/or molecular confirmation on an EcM association.
The /sowerbyella lineage comprises the genus Sowerbyella that consists of 14 species (Yao and Spooner 2006). Members of the genus are typically found on the forest floor with EcM hosts. The rooting habit of the fruiting bodies, the fact that no member of the genus has been grown in axenic culture, and the isotopic signatures of some species (Hobbie et al. 2001) suggest that this genus may be EcM. However, there is still is no good molecular or anatomical data to show the EcM habit in this group. Hansen et al. (2013) resolved Sowerbyella on a branch among non-EcM relatives (e.g., Aleuria, Lasiobolidium) and suggested that Sowerbyella may not be EcM.
The multi-locus phylogenetic analysis of Sánchez-García et al. (2014) that separated /porpoloma from /tricholoma also proposed Albomagister as a segregate genus that is phylogenetically distinct from other Tricholomataceae. Albomagister was hypothesized to be EcM, because species in this genus fruit on the forest floor in association with EcM Fagales and Pinaceae and have δ15N and δ13C isotopic signatures that are similar to some other EcM fungi (e.g., members of the /catathelasma lineage) (Birkebak et al. 2013). It is possible that Albomagister represents another independent EcM lineage but root tips have never been sampled to test this hypothesis further.
6.7 New Additions of Genera Confirmed as Ectomycorrhizal
The genera of several EcM fungal lineages have been recently revised, resulting mostly in the splitting of large and heterogeneous genera into smaller groups. In the /sebacina lineage, the early diverging genus Helvellosebacina was separated from the rest of Sebacina whereas Tremellodendron was merged into Sebacina (Oberwinkler et al. 2014). The genus Tremelloscypha was resolved as the sister lineage to Sebacina and Helvellosebacina (but see Tedersoo et al. (2014b)), and all members of all three genera form a monophyletic group of EcM taxa. Oberwinkler et al. (2014) placed all of the non-mycorrhizal taxa in segregate genera, including Chaetospermum, Craterocolla, Globulisebacina (comprising Efibulobasidium rolleyi), and Paulisebacina (comprising Sebacina allantoidea).
The genus Psathyloma was provisionally included in the /hebeloma-alnicola lineage by Tedersoo and Smith (2013), but it was officially described only recently (Soop et al. 2016). It comprised three species, viz., P. leucocarpum and P. catervatim in New Zealand and Tasmania, and a third undescribed species from Argentina (root tip: JX316416). All known sequences and specimens of Psathyloma are known from Southern Hemisphere Nothofagus forests. In the analysis of Soop et al. (2016), the genus Psathyloma was resolved as the sister lineage to other taxa in the /hebeloma-alnicola lineage, suggesting the possibility of an ancient divergence between Psathyloma and all other genera in this group.
The /boletus lineage has been a subject to explosive radiation of descriptions of novel genera, several of which turn out to be non-monophyletic after addition of new taxa or information from other genes. The recent additions include Alessioporus, Pulchroboletus (Gelardi et al. 2014a), Baorangia, Lanmaoa, Parvixerocomus, Rugiboletus (Wu et al. 2015), Binderoboletus, Guyanaboletus, Singerocomus (Henkel et al. 2016), Butyriboletus (Arora and Frank 2014), Caloboletus (Vizzini 2014a), Castellanea, Costatisporus, Jimtrappea (Smith et al. 2015), Cupreoboletus (Gelardi et al. 2015a), Crocinoboletus (Zeng et al. 2014), Cyanoboletus (Gelardi et al. 2014b), Exsudoporus (Vizzini 2014b), Imleria (Vizzini 2014d), Hourangia (Zhu et al. 2015), Neoboletus (Vizzini 2014c), Nigroboletus (Gelardi et al. 2015b), Pseudoaustroboletus (Li et al. 2014), and Rubroboletus (Zhao et al. 2014).
6.8 Notes on the /Elaphomyces Lineage
The monophyly of the /elaphomyces lineage and the genus Elaphomyces was recently questioned by Buyck et al. (2016). Although we agree that this group warrants further taxonomic and phylogenetic research, we disagree with the suggestion that the African sequences published in Tedersoo et al. (2011b) are erroneous. We also disagree with the weak evidence that was presented for the polyphyly of this group of sequestrate hypogeous fungi. Re-evaluating the sequence data revealed that BLASTn results were meaningful only when conservative parameters (word size = 7, match score = 1, mismatch score = 3, gap opening cost = 5, gap extension cost = 2) but not MegaBLAST parameters were chosen. Buyck et al. (2016) also used only the ITS region for analysis and selected a specimen from another subclass (Chaetothyriomycetidae) as outgroup. Since the sequences of multiple clades within the /elaphomyces lineage are not alignable due to extremely high ITS sequence divergence (particularly among some undescribed tropical taxa), any phylogenetic analyses are likely to generate spurious results. We consider this study to be misleading and insufficient to suggest the polyphyly of Elaphomycetaceae or the /elaphomyces lineage. Here we do not make any changes in regard to the /elaphomyces lineage, but we do recommend caution when assigning sequences to the /elaphomyces lineage based on BLASTn searches.
6.9 Saprotrophic, Facultatively Biotrophic Phlebopus
In Tedersoo et al. (2010, p. 243), we discussed the mycorrhizal status of Phlebopus and considered this genus to be non-EcM but biotrophic. Phlebopus spp. readily form fruit bodies without any EcM host plants in sterile and nonsterile media and in natural conditions (Ji et al. 2011; Zhang et al. 2015; Kumla et al. 2016). In nature, Phlebopus spp. grow superficially and colonize the epidermal cells of AM and rarely EcM plants and associate with scale insects that form root galls in these roots (Zhang et al. 2015 and references therein). In axenic and synthesis trials in sterile and nonsterile substrate, Phlebopus spp. are reported to form ectomycorrhizal structures with EcM Australian Acacia spp. (Thoen and Ducouso 1989) and Pinus kesiya (Kumla et al. 2016). Although the illustrations of synthesized EcM structures are convincing in the latter study, we cannot accept Phlebopus as ectomycorrhizal because these associations are lacking or extremely rare in natural conditions (Zhang et al. 2015). We interpret the root-associated habit of Phlebopus as biotrophic but both non-mycorrhizal and non-parasitic, because the inoculated plants show no signs of decline (Kumla et al. 2016). The biotrophic associations with both roots and scale insects are likely facultative, because Phlebopus spp. are able to complete their life cycle saprotrophically without any of these interactions.
6.10 Recognition of EcM Fungal Lineages
Based on the criteria in Sect. 6.2, we propose specific criteria for separation of EcM fungal lineages from related non-EcM groups (Table 6.1) using the ITS2 subregion and full ITS. For ITS2 and full ITS, respectively, 45 (53%) and 60 (70%) lineages could be reliably delimited based on the BLASTn score to query sequence length (S/L) ratio alone because of a significant phylogenetic gap between EcM and closely related non-EcM groups. One quarter of lineages exhibited a small range of S/L values, where trophic assignment is unambiguous. In these cases, assignment of individual lineages should be sought for support by manual BLASTn queries and/or phylogenetic analyses for greater reliability. In general, placement tended to be relatively more ambiguous for the most diverse EcM groups such as the /russula-lactarius, /inocybe, /clavulina, and /boletus lineages but not in the /tomentella-thelephora and /cortinarius lineages. Phylogenetic analyses suggested ambiguity in cases where the non-EcM outgroup(s) was separated by a relatively short stem (e.g., /tricholoma: Sánchez-García et al. 2014), or the outgroup had a low rate of ITS evolution (e.g., /inocybe: Ryberg et al. 2010), or there in rapidly evolving clades within the EcM lineages (e.g., /clavulina: Kennedy et al. 2012; /boletus: Nuhn et al. 2013). Except for /hysterangium, /inocybe, and /clavulina, <2% of OTUs across the lineages of ambiguously delimited groups fell into the uncertain range of S/L values, indicating the overall rate for correct placement at 97–98%.
6.11 Conclusions
With the addition of the /leotia, /porpoloma, /endogone3, and /phaeocollybia lineages to information from a previous review (Tedersoo and Smith 2013), the number of EcM fungal lineages has now grown to 82–86 separate groups comprising 279–284 genera. The rate of discovery of novel EcM lineages is notably declining because the most common groups have been already described. This is due to a huge increase in the number of in situ molecular identification studies of EcM fungal communities on roots as compared to a decade ago. However, the number of profound EcM community studies tends to decline in recent years, because most laboratories have switched to HTS-based identification of EcM fungi directly from bulked root and soil samples. Since EcM fungi naturally co-occur with many other fungal and eukaryote groups, it is impossible to verify the EcM habit from these types of studies. Our overview about the parameters of semiautomatic EcM lineage recognition should enable accurate trophic assignment of 95–99% of fungal OTUs to EcM and non-EcM categories. Further developments in this field should include development and automatized application of taxon-specific sequence similarity thresholds for taxa by using expert molecular taxonomic knowledge. In the future, it will also be important to use additional, phylogenetically or functionally informative loci for HTS-based approaches beyond ITS sequencing. It should also be possible in the future to automatically assign traits and functions to the EcM fungi based on a combination of the taxonomy and what is known about reference taxa. Much has yet to be done to incorporate information about functional genes of taxa obtained from genomics studies and using probabilistic approaches rather than binary (presence/absence) functional assignments.
References
Argüelles-Moyao A, Garibay-Orijel R, Márquez-Valdelamar LM, Arellano-Torres E (2017) Clavulina-Membranomyces is the most important lineage within the highly diverse ectomycorrhizal fungal community of Abies religiosa. Mycorrhiza 27:53–65
Arora D, Frank JL (2014) Clarifying the butter Boletes: a new genus, Butyriboletus, is established to accommodate Boletus sect. Appendiculati, and six new species are described. Mycologia 106:464–480
Baral H-O, Galan R, Platas G, Tena R (2013) Phaeohelotium undulatum comb. nov. and Phaeoh. succineoguttulatum sp. nov., two segregates of the Discinella terrestris aggregate found under Eucalyptus in Spain: taxonomy, molecular biology, ecology and distribution. Mycosystema 32:386–428
Berbee ML, Wong EY, Tsui CK (2016) Phylogenetic evidence places the coralloid jelly fungus Tremellodendropsis tuberosa (Tremellodendropsidales) among early diverging Agaricomycetes. Mycol Prog 15:939–946
Berch SM, Castellano MA (1986) Sporulation of Endogone pisiformis in axenic and monoxenic culture. Mycologia 78:292–295
Berch SM, Fortin JA (1983a) Endogone pisiformis: axenic culture and associations with Sphagnum, Pinus sylvestris, Allium cepa and Allium porrum. Can J Bot 61:899–905
Berch SM, Fortin JA (1983b) Germination of zygospores of Endogone pisiformis. Mycologia 75:328–332
Birkebak JM, Mayor JR, Ryberg KM, Matheny PB (2013) A systematic, morphological and ecological overview of the Clavariaceae (Agaricales). Mycologia 105:896–911
Buyck B, Hosaka K, Masi S, Hofstetter V (2016) Molecular analyses of first collections of Elaphomyces Nees (Elaphomycetaceae, Eurotiales, Ascomycota) from Africa and Madagascar indicate that the current concept of Elaphomyces is polyphyletic. Cryptogam Mycol 37:3–14
Coimbra VRM, Gibertoni TB, Wartchow F (2012) Phaeocollybia nigripes (Agaricomycetes), a new species from Brazil. Mycotaxon 120:171–179
Fernández NV, Marchelli P, Fontenla SB (2013) Ectomycorrhizas naturally established in Nothofagus nervosa seedlings under different cultivation practices in a forest nursery. Microb Ecol 66:581–592
Field KJ, Rimington WR, Bidartondo MI, Allinson KE, Beerling DJ, Cameron DD, Duckett JG, Leake JR, Pressel S (2015) First evidence of mutualism between ancient plant lineages (Haplomitropsida liverworts) and Mucoromycotina fungi and its response to simulated Paleozoic changes in atmospheric CO2. New Phytol 205:743–756
Gelardi M, Simonini G, Ercole E, Vizzini A (2014a) Alessioporus and Pulchroboletus gen. nov. (Boletaceae, Boletineae), two novel genera to accommodate Xerocomus ichnusanus and X. roseoalbidus from European Mediterranean basin: molecular and morphological evidence. Mycologia 106:1168–1187
Gelardi M, Vizzini A, Simonini G (2014b) Cyanoboletus. Index Fungorum 176:1
Gelardi M, Simonini G, Ercole E, Davoli P, Vizzini A (2015a) Cupreoboletus (Boletaceae, Boletineae), a new monotypic genus segregated from Boletus sect. Luridi to reassign the Mediterranean species B. poikilochromus. Mycologia 107:1254–1269
Gelardi M, Vizzini A, Ercole E, Horak E, Ming Z, Li TH (2015b) Circumscription and taxonomic arrangement of Nigroboletus roseonigrescens Gen. et Sp. nov., a new member of Boletaceae from tropical South–Eastern China. PLoS One 10:e0134295
Hansen K, Perry BA, Dranginis AW, Pfister DH (2013) A phylogeny of the highly diverse cup-fungus family Pyronemataceae (Pezizomycetes, Ascomycota) clarifies relationships and evolution of selected life history traits. Mol Phylogenet Evol 67:311–335
Henkel TW, Obase K, Husbands D, Uehling JK, Bonito G, Aime MC, Smith ME (2016) New Boletaceae taxa from Guyana: Binderoboletus segoi gen. and sp. nov., Guyanaporus albipodus gen. and sp. nov., Singerocomus rubriflavus gen. and sp. nov., and a new combination for Xerocomus inundabilis. Mycologia 108:157–173
Hirose D, Degawa Y, Yamamoto K, Yamada A (2014) Sphaerocreas pubescens is a member of the Mucoromycotina closely related to fungi associated with liverworts and hornworts. Mycoscience 55:221–226
Hobbie EA, Weber NS, Trappe JM (2001) Mycorrhizal vs. saprotrophic status of fungi: the isotopic evidence. New Phytol 150:601–610
Jabaji-Hare SH, Charest PM (1987) Ultrastructural and cytochemical observations on the somatic phase of Endogone pisiformis (Endogonaceae). Mycologia 79:433–444
Ji KP, Cao Y, Zhang CX, He MX, Liu J, Wang WB, Wang Y (2011) Cultivation of Phlebopus portentosus in southern China. Mycol Prog 10:293–300
Kennedy PG, Matheny PB, Ryberg KM, Henkel TW, Uehling JK, Smith ME (2012) Scaling up: examining the macroecology of ectomycorrhizal fungi. Mol Ecol 21:4151–4154
Kohler A, Kuo A, Nagy LG, Morin E, Barry KW, Buscot F, Canbäck B, Tunlid A, Grigoriev IV, Hibbett DS, Martin F (2015) Convergent losses of decay mechanisms and rapid turnover of symbiosis genes in mycorrhizal mutualisms. Nat Genet 47:410–415
Kõljalg U, Tedersoo L, Nilsson RH, Abarenkov K (2016) Digital identifiers for fungal species. Science 352:1182–1183
Kühdorf K, Münzenberger B, Begerow D, Gomez-Laurito J, Hüttl RF (2015) Leotia cf. lubrica forms arbutoid mycorrhiza with Comarostaphylis arbutoides (Ericaceae). Mycorrhiza 25:109–120
Kumla J, Hobbie EA, Suwannarach N, Lumyong S (2016) The ectomycorrhizal status of a tropical black bolete, Phlebopus portentosus, assessed using mycorrhizal synthesis and isotopic analysis. Mycorrhiza 26:333–343
Li YC, Li F, Zeng NK, Cui YY, Yang ZL (2014) A new genus Pseudoaustroboletus (Boletaceae, Boletales) from Asia as inferred from molecular and morphological data. Mycol Prog 13:1207–1216
McGee PA (1996) The Australian zygomycetous mycorrhizal fungi: the genus Densospora gen. nov. Aust Syst Bot 9:329–336
Nguyen NH, Song Z, Bates ST, Branco S, Tedersoo L, Menke J, Schilling JS, Kennedy PG (2016) FUNGuild: an open annotation tool for parsing fungal community data sets by ecological guild. Fungal Ecol 20:241–248
Nouhra E, Urcelay C, Longo S, Tedersoo L (2013) Ectomycorrhizal fungal communities associated to Nothofagus species in Northern Patagonia. Mycorrhiza 23:487–496
Nuhn M, Binder M, Taylor AFS, Halling RE, Hibbett DS (2013) Phylogenetic overview of the Boletineae. Fungal Biol 117:479–511
Obase K, Douhan G, Matsuda Y, Smith ME (2016) Revisiting phylogenetic diversity and cryptic species of Cenococcum geophilum sensu lato. Mycorrhiza 26:529–540
Oberwinkler F, Riess K, Bauer R, Garnica S (2014) Morphology and molecules: the Sebacinales, a case study. Mycol Prog 13:445–470
Orchard S, Hilton S, Bending GD, Dickie IA, Standish RJ, Gleeson DB, Jeffery RP, Powell JR, Walker C, Bass D, Monk J (2017) Fine endophytes (Glomus tenue) are related to Mucoromycotina, not Glomeromycota. New Phytol 213:481–486
Porras-Alfaro A, Liu K-L, Kuske CR, Xie G (2014) From genus to phylum: large subunit and internal transcribed spacer rRNA operon regions show similar classification accuracies influenced by database composition. Appl Environ Microbiol 80:829–840
Redhead S, Malloch DW (1985) The genus Phaeocollybia (Agaricales) in eastern Canada and its biological status. Can J Bot 64:1249–1254
Rinaldi AC, Comandini O, Kuyper TW (2008) Ectomycorrhizal fungal diversity: separating the wheat from the chaff. Fungal Divers 33:1–45
Ryberg M, Larsson E, Jacobsson S (2010) An evolutionary perspective on morphological and ecological characters in the mushroom family Inocybaceae. Mol Phylogenet Evol 55:431–442
Sánchez-García M, Matheny PB, Palfner G, Lodge DJ (2014) Deconstructing the Tricholomataceae (Agaricales) and introduction of the new genera Albomagister, Corneriella, Pogonoloma and Pseudotricholoma. Taxon 63:993–1007
Sánchez-García M, Henkel TW, Aime MC, Smith ME, Matheny PB (2016) Guyanagarika, a new ectomycorrhizal genus of Agaricales from the Neotropics. Fungal Biol 120:1540–1553
Smith ME, Amses KR, Elliott TF, Obase K, Aime MC, Henkel TW (2015) New sequestrate fungi from Guyana: Jimtrappea guyanensis gen., sp. nov., Castellanea pakaraimophila gen., sp. nov., and Costatisporus cyanescens gen., sp. nov. (Boletaceae, Boletales). IMA Fungus 6:297–317
Soop K, Dima B, Szarkándi JG, Cooper J, Papp T, Vágvölgyi C, Nagy LG (2016) Psathyloma, a new genus in Hymenogastraceae described from New Zealand. Mycologia 108:397–404
Spatafora JW, Owensby CA, Douhan GW, Boehm EWA, Schoch CL (2012) Phylogenetic placement of the ectomycorrhizal Cenococcum in Gloniaceae. Mycologia 104:758–765
Spatafora JW, Chang Y, Benny GL, Lazarus K, Smith ME, Berbee ML, Bonito G, Corradi N, Grigoriev I, Gryganskyi A, James TY, Stajich J (2016) A phylum-level phylogenetic classification of zygomycete fungi based on genome-scale data. Mycologia 108:1028–1046
Tedersoo L, Lindahl B (2016) Fungal identification biases in microbiome projects. Environ Microbiol Rep 8:774–779
Tedersoo L, Nilsson RH (2016) Molecular identification of fungi. In: Martin F (ed) Molecular mycorrhizal symbiosis. Wiley, London, pp 301–322
Tedersoo L, Smith ME (2013) Lineages of ectomycorrhizal fungi revisited: foraging strategies and novel lineages revealed by sequences from belowground. Fungal Biol Rev 27:83–99
Tedersoo L, Jairus T, Horton BM, Abarenkov K, Suvi T, Saar I, Kõljalg U (2008) Strong host preference of ectomycorrhizal fungi in a Tasmanian wet sclerophyll forest as revealed by DNA barcoding and taxon-specific primers. New Phytol 180:479–490
Tedersoo L, May TW, Smith ME (2010) Ectomycorrhizal lifestyle in fungi: global diversity, distribution, and evolution of phylogenetic lineages. Mycorrhiza 20:217–263
Tedersoo L, Abarenkov K, Nilsson RH, Schüβler A, Grelet G-A, Kohout P, Oja J, Bonito GM, Veldre V, Jairus T, Ryberg M, Larsson K-H, Kõljalg U (2011a) Tidying up International Nucleotide Sequence Databases: ecological, geographical and sequence quality annotation of ITS sequences of mycorrhizal fungi. PLoS One 6:e24940
Tedersoo L, Bahram M, Jairus T, Bechem E, Chinoya S, Mpumba R, Leal M, Randrianjohany E, Razafimandimbison S, Sadam A, Naadel T, Kõljalg U (2011b) Spatial structure and the effects of host and soil environments on communities of ectomycorrhizal fungi in wooded savannas and rain forests of Continental Africa and Madagascar. Mol Ecol 20:3071–3080
Tedersoo L, Bahram M, Põlme S, Kõljalg U, Yorou NS, Wijesundera R, Villarreal-Ruiz L, Vasco-Palacios A, Quang Thu P, Suija A, Smith ME, Sharp C, Saluveer E, Saitta A, Ratkowsky D, Pritsch K, Riit T, Põldmaa K, Piepenbring M, Phosri C, Peterson M, Parts K, Pärtel K, Otsing E, Nouhra E, Njouonkou AL, Nilsson RH, Morgado LN, Mayor J, May TW, Kohout P, Hosaka K, Hiiesalu I, Henkel TW, Harend H, Guo L, Greslebin A, Grelet G, Geml J, Gates G, Dunstan W, Dunk C, Drenkhan R, Dearnaley J, De Kesel A, Dang T, Chen X, Buegger F, Brearley FQ, Bonito G, Anslan S, Abell S, Abarenkov K (2014a) Global diversity and geography of soil fungi. Science 346:1078
Tedersoo L, Bahram M, Ryberg M, Otsing E, Kõljalg U, Abarenkov K (2014b) Global biogeography of the ectomycorrhizal/sebacina lineage (Fungi, Sebacinales) as revealed from comparative phylogenetics analyses. Mol Ecol 23:4168–4183
Tedersoo L, Anslan S, Bahram M, Põlme S, Riit T, Liiv I, Kõljalg U, Kisand V, Nilsson RH, Bork P, Hildebrand F, Abarenkov K (2015a) Shotgun metagenomes and multiple primer pair-barcode combinations of amplicons reveal biases in metabarcoding analyses of fungi. MycoKeys 10:1–43
Tedersoo L, Bahram M, Põlme S, Anslan S, Riit T, Kõljalg U, Nilsson RH, Hildebrand F, Abarenkov K (2015b) Response to comment on “global diversity and geography of soil fungi”: analytical biases in microbial diversity studies. Science 359:936
Tedersoo L, Bahram M, Cajthaml T, Põlme S, Hiiesalu I, Anslan S, Harend H, Buegger F, Pritsch K, Koricheva J, Abarenkov K (2016a) Tree diversity and species identity effects on soil fungi, protists and animals are context-dependent. ISME J 10:346–362
Tedersoo L, Liiv I, Kivistik PA, Anslan S, Kõljalg U, Bahram M (2016b) Genomics and metagenomics technologies to recover ribosomal DNA and single-copy genes from old fruitbody and ectomycorrhiza specimens. MycoKeys 13:1–20
Thoen D, Ducouso M (1989) Mycorrhizal habit and sclerogenesis of Phlebopus sudanicus (Gyrodontaceae) in Senegal. Agric Ecosyst Environ 28:519–523
Truong C, Mujic A, Healy R, Kuhar F, Furci G, Torres D, Niskanen T, Sandoval-Leiva P, Fernandez N, Escobar J, Moretto A, Palfner G, Pfister D, Nohra E, Swenie R, Sanchez-Garcia M, Matheny PB, Smith ME (2017). How to know the fungi: combining field inventories and DNA-barcoding to document fungal diversity. New Phytol. doi:10.1111/nph.14509
Vizzini A (2014a) Nomenclatural novelties. Index Fungorum 146:1–2
Vizzini A (2014b) Nomenclatural novelties. Index Fungorum 183:1
Vizzini A (2014c) Nomenclatural novelties. Index Fungorum 192:1
Vizzini A (2014d) Nomenclatural novelties. Index Fungorum 147:1
Vizzini A, Consiglio G, Ercole E, Setti L (2016) Pseudoporpoloma, a new genus for Agaricus pes-caprae (Agaricales, Tricholomataceae). Phytotaxa 243:271–280
Warcup JH (1985) Ectomycorrhiza formation by Glomus tubiforme. New Phytol 99:267–272
Warcup JH (1990a) Occurrence of ectomycorrhizal and saprophytic discomycetes after a wild fire in an eucalypt forest. Mycol Res 94:1065–1069
Warcup JH (1990b) Taxonomy, culture and mycorrhizal associations of some zygosporic Endogonaceae. Mycol Res 94:173–178
Wu G, Zhao K, Li YC, Zeng NK, Feng B, Halling RE, Yang ZL (2015) Four new genera of the fungal family Boletaceae. Fungal Divers 2015:1–24
Yamamoto K, Degawa Y, Hirose D, Fukuda M, Yamada A (2015) Morphology and phylogeny of four Endogone species and Sphaerocreas pubescens collected in Japan. Mycol Prog 14:86
Yamamoto K, Endo N, Degawa Y, Fukuda M, Yamada A (2017) First detection of Endogone ectomycorrhizas in natural oak forests. Mycorrhiza 27:295–301
Yao YJ, Spooner BM (2006) Species of Sowerbyella in the British Isles, with validation of Pseudoombrophila sect. Nannfeldtiella (Pezizales). Fungal Divers 22:267–279
Zeller B, Bréchet C, Maurice J-C, Le Tacon F (2007) 13C and 15N isotopic fractionation in trees, soils and fungi in a natural forest stand and Norway spruce plantation. Ann For Sci 64:419–429
Zeng NK, Wu G, Li YC, Liang ZQ, Yang ZL (2014) Crocinoboletus, a new genus of Boletaceae (Boletales) with unusual boletocrocin polyene pigments. Phytotaxa 175:133–140
Zhang CX, He MX, Cao Y, Liu J, Gao F, Wang WB, Ji KP, Shao SC, Wang Y (2015) Fungus-insect gall of Phlebopus portentosus. Mycologia 107:12–20
Zhao K, Wu K, Yang ZL (2014) A new genus, Rubroboletus, to accommodate Boletus sinicus and its allies. Phytotaxa 188:61–77
Zhu XT, Wu G, Zhao K, Halling RE, Yang ZL (2015) Hourangia, a new genus of Boletaceae to accommodate Xerocomus cheoi and its allied species. Mycol Prog 14:1–10
Acknowledgments
We thank K. Yamamoto and the non-anonymous referees K. Kühdorf and M. Sanchez-Garcia for critical comments on the manuscript. L.T. acknowledges funding from the Estonian Science Foundation, PUT1399/MOBERC1. Funding for M. E. Smith was provided in part by the US National Science Foundation grants DEB-1354802 and DEB-1441677 and by the University of Florida’s Institute for Food and Agricultural Sciences (IFAS).
Author information
Authors and Affiliations
Corresponding author
Editor information
Editors and Affiliations
Rights and permissions
Copyright information
© 2017 Springer International Publishing AG
About this chapter
Cite this chapter
Tedersoo, L., Smith, M.E. (2017). Ectomycorrhizal Fungal Lineages: Detection of Four New Groups and Notes on Consistent Recognition of Ectomycorrhizal Taxa in High-Throughput Sequencing Studies. In: Tedersoo, L. (eds) Biogeography of Mycorrhizal Symbiosis. Ecological Studies, vol 230. Springer, Cham. https://doi.org/10.1007/978-3-319-56363-3_6
Download citation
DOI: https://doi.org/10.1007/978-3-319-56363-3_6
Published:
Publisher Name: Springer, Cham
Print ISBN: 978-3-319-56362-6
Online ISBN: 978-3-319-56363-3
eBook Packages: Biomedical and Life SciencesBiomedical and Life Sciences (R0)