Abstract
Imaging of infection in the CNS has been handled using cross-sectional imaging for more than two decades now resulting in a large array of descriptive diagnostic criteria, capable, in most circumstances of narrowing the differential diagnosis, detecting life-threatening complications and establishing baseline for assessment of treatment response. Limitations however exist, and in many circumstances, both cross-sectional imaging and nonspecific molecular imaging, such as 18F-FDG, fail to establish a diagnosis. The availability of pathogen-specific imaging agents/ligands would have a great effect on the management of patients with CNS infection. Besides early diagnosis, avoidance of diagnostic brain biopsies can have significant effect on the mortality and morbidity of patients.
You have full access to this open access chapter, Download chapter PDF
Similar content being viewed by others
Keywords
- Thymidine Kinase
- Simian Immunodeficiency Virus
- Central Nervous System Infection
- Mouse Hepatitis Virus
- Experimental Cerebral Malaria
These keywords were added by machine and not by the authors. This process is experimental and the keywords may be updated as the learning algorithm improves.
8.1 Introduction
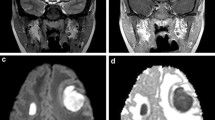
Since the advent of cross-sectional magnetic resonance (MR) and computed tomography (CT) imaging techniques, the diagnosis and characterization of various intracranial infectious processes has advanced significantly. Specific imaging characteristics for different pathogens were thoroughly described and now represent the first step in the management and follow-up of those patients. Structural cross-sectional techniques , however, reach their limits when questions of molecular nature are asked, such as differentiating active infection from sterile inflammation and/or immune response or from other etiologies such as primary or metastatic malignancies (Figure 8.1). Molecular imaging, on the other hand, has the potential of transcending the limitations of structural imaging, providing molecular and cellular level information. Multiple examples of preclinical central nervous system (CNS) imaging applications in the setting of viral, bacterial, parasitic, and fungal infections can be found in the literature. Clinical and human translational applications however remain limited in number and in scope.
Optical imaging and PET/SPECT imaging are the two most commonly used molecular imaging modalities in animal models of CNS infection. While only a few studies have attempted targeting the unmodified pathogen directly, tracking genetically modified pathogens in animal models, generally using optical imaging, remains the main approach. There are also multiple studies describing host responses to infection such as activation of microglia or other components of the immune system. Among those approaches, direct targeting of the pathogen is probably of most clinical significance and would be most interesting to translate.
8.2 Targeting the Pathogen
Targeting the pathogen is limited by lack of pathogen-specific imaging ligands . Another limitation inherent to the CNS is the presence of the blood–brain barrier limiting brain access to few molecules with special characteristics (e.g., small size, appropriate lipophilicity). Even though the BBB can be disrupted in infectious processes, this is not always the case. In fact, with indolent less aggressive pathogens, the BBB can remain intact or minimally breached. In theory, directly imaging the infectious agent can be performed using ligands incorporated only by the pathogen but not by eukaryotic cells or through targeting of pathogen enzymes with labeled substrates. One example of the latter approach is imaging herpetic (HSV-1 ) encephalitis using 18F-FHPG, a fluorinated derivative of ganciclovir, which is selectively phosphorylated by the HSV thymidine kinase (Tk) enzyme and subsequently trapped within infected cells [1]. Selective uptake of the ligand is then visualized using PET imaging of rats that received intranasal injection of HSV-1 compared to sham animals. This approach however is limited to pathogens that possess the Tk enzyme, but more importantly by the inability of Tk substrates such as FHPG, FHBG, and FIAU to cross an intact BBB . Even when the BBB is disrupted, the substrate can “leak” resulting in nonspecific accumulation. A well-designed experiment by Buursma et al. used a freezing injury model causing sterile disruption of the BBB ; even though 18F-FHPG accumulated initially, it washed out quickly suggesting that the FHPG uptake in actual infections is due to irreversible trapping of the ligand in phosphorylated form within the infected cells [1].
Another recently described ligand, 2-[18F]-fluorodeoxysorbitol (18F-FDS) , depends on its selective uptake by bacteria, in this case, the Enterobacteriaceae family of Gram-negative bacteria. Sorbitol is not taken by eukaryotic cells and was found to be readily incorporated in Gram-negative but not Gram-positive bacterial cells [2]. 18F-FDS was previously synthesized through chemical reduction of the widely available ligand, 18F-2-fluoro-deoxy-d-glucose (18F-FDG) [3]. Even though originally designed to image brain tumors [3], a significantly higher uptake of 18F-FDS was seen in the E. coli-infected brain versus brain tumor or normal brain, suggesting specific uptake and retention by the bacteria (Figure 8.2) [2]. This is rendered even more sensitive by the demonstrated lack of background uptake in the normal brain. Even though FDS can only detect infections due to the Enterobacteriaceae family , it can be greatly used in the right clinical setting, providing localization of the infection, differentiation of active infection from host inflammatory reaction, and evaluation of response to treatment. Development of pathogen-specific ligands with wider detection range and low background uptake is the ultimate goal.
18F-FDS PET signal in an E. coli-infected brain (top row), normal brain (middle row), and brain tumor tissues (lower row). Uptake of the ligand is only noted in association with Gram-negative infection suggesting specific uptake by the pathogen. Adapted from Weinstein and Ordonez et al. [2]
8.3 Genetically Modified Pathogens and Reporter Mice
Much more commonly seen than direct pathogen detection in the brain is molecular imaging using genetically modified pathogens that are traceable through a reporter gene mechanism. A common approach is the use of recombinant viruses engineered to express a luciferase enzyme which allows performing longitudinal imaging, accurately determining the site(s) of infection, describing the temporal systemic dissemination of the virus, and eventually quantifying viral titers in various organs. A major limitation however is the inherent potential for attenuation of viral replication and/or loss of virulence of the pathogen [4,5,6,7,8]. Both latter factors thus need to be carefully evaluated prior to starting imaging experiments. Early examples included developing recombinant viruses that expressed various luciferase reporter proteins such as HSV-1 [6], vaccinia virus [9], and the mouse hepatitis virus (MHV) , a member of the Coronavirus family [10]. An interesting finding in the paper by Raaben et al. is the detection of the protective role of type I interferon in MHV infection , with widespread viral dissemination, elucidated through bioluminescence, seen in mice lacking a functional type I interferon response and reversal of the process through the administration of type I interferons to the mice [10]. This is an example of molecular imaging providing insight into potential therapeutic applications. More recent examples include the use of recombinant murine gammaherpesvirus 68 (MHV-68) expressing the firefly luciferase (Fluc) to monitor virus progression after CNS infection [11] and of recombinant Dengue virus , for real-time evaluation of replication kinetics in the brain of infected mice [12]. Western equine encephalitis virus (WEEV; Alphavirus) , a mosquito-borne virus that can cause severe encephalitis in humans, was also evaluated in a recombinant form (expressing Fluc) in intranasally infected mice [13].
The genetically engineered reporter gene approach however is not limited to viruses and has been used with other pathogens such as parasites. A genetically modified and fully virulent African IL1392 Trypanosoma vivax strain that stably expresses Fluc was monitored in vivo in the brain of infected immunocompetent mice. Light emitted by the brain increased slightly up till day 20 post-infection (<400-fold the background) then rose substantially by day 25, reaching up to 2000-fold the background, thus providing evidence of parenchymal infiltration [14]. Other examples of reporter gene use include imaging of Toxoplasma gondii encephalitis with spatiotemporal demonstration of recrudescence of the infection from the CNS of immunocompromised mice [15].
Virulent recombinant pathogens can also be used to assess responsiveness to treatment. For example, a Streptococcus pneumoniae strain modified by the integration of a luminescent lux operon was used to determine responsiveness of the infection to daptomycin, an alternative antibiotic, eventually found to be as effective as vancomycin [16]. Similarly, the use of luciferase-expressing Trypanosoma brucei brucei allowed the identification of pathogen entry into the brain (using multi-photon microscopy ) in early stages and determination of their responsiveness to various drugs (bioluminescent parasites were permanently eliminated by treatment with melarsoprol and DB829 but not with diminazene). The interesting finding in this paper is that the investigators did not have to wait for the clinical development of the encephalitic stage of disease which could take as late as 180 days after the initial stage of the infection. Instead, by using serial noninvasive bioluminescence imaging, they reduced the time needed to assess drug efficacy by two-thirds [17].
An alternative model to reporter gene-expressing pathogens is to genetically modify the animal model, usually mice. For example, Luker et al. developed a transgenic reporter mouse that uses the promoter from HSV-1 thymidine kinase to control expression of Fluc [6]. Exposure to HSV-1 (three different strains tested) would then activate the expression of Fluc with bioluminescence increasing in proportion to increasing input titers of the virus. This study however did not detect brain involvement with HSV-1. More complex models such as the transgenic reporter mouse strain that expresses Fluc under the regulatory control of a concatenated Gal4 promoter [18] and the transgenic reporter mouse in which luciferase expression is driven by the nuclear factor B (NF-B)-dependent portion of the human immunodeficiency virus-1 long terminal repeat (HIV-1 LTR) [19] allowed the visualization of brain luciferase expression in response to adenovirus infection [18] and LPS intraperitoneal injection [19], respectively.
A particularly interesting example combines the two approaches (reporter gene and reporter mouse ), using Streptococcus pneumoniae strain engineered for bioluminescence (lux) in a transgenic mouse model containing an inducible Fluc reporter gene under transcriptional control of the mouse glial fibrillary acidic protein (GFAP) promoter . The two reporters were then separated based on the different spectra of light emission and substrate requirements for lux and Fluc, using a highly sensitive in vivo imaging system. This approach allowed the monitoring of disease progression and response to treatment on one hand while documenting brain damage at the same time [20].
8.4 Imaging the Host
An indirect way of imaging the sequelae of intracranial infection is by imaging the host reaction . Perhaps the most commonly used approach in this setting is imaging infection-associated neuroinflammation . This can be achieved through targeting of the translocator protein (TSPO) , an outer mitochondrial membrane receptor that gets upregulated in activated microglia, astrocytes, and blood- derived macrophages in response to CNS central nervous system injury. The classical ligand for TSPO has always been 11C-PK11195; however a multitude of newer (second-generation) ligands designed to overcome the limitations of PK11195 have been developed over the last couple of decades [21]. Using two of the second-generation ligands, 11C-DPA-713 and 18F-DPA-714, along with the prototype ligand 11C-PK11195 [22], neuroinflammation in the setting of HSV encephalitis could be imaged, with 11C-DPA713 showing the lowest nonspecific uptake, suggesting it may be the most suitable for visualizing mild inflammation. Other pathogens in which neuroinflammation was visualized using TSPO ligands include simian immunodeficiency virus (SIV) in macaques [21] and Taenia solium (neurocysticercosis) in patients [23]. In the latter study, perilesional edema surrounding calcified lesions was visualized using 11C-PBR28 suggesting resurgence of inflammation despite the chronicity of the infectious process.
Major limitations of TSPO imaging however exist. The main limitation is its inability to specifically visualize infection with ligand accumulation readily seen with sterile inflammation such as that induced by injection of quinolinic acid in the rat striatum [24] (Figure. 8.3). Another main limitation in human translation of TSPO imaging is a recently described polymorphism (rs6971) located in exon 4 of the TSPO gene which results in nonconservative amino acid substitution at position 147 from alanine to threonine (Ala147Thr ) in the fifth transmembrane domain of the TSPO protein [25]. This polymorphism results in three binding levels: high, medium, and low binding. To account for the variability, in most cases, genotyping of subjects prior to quantitative assessments of TSPO upregulation using PET is necessary. Another problem with TSPO imaging in non-focal brain infections such as neuro-HIV is the lack of a reference region due to diffuse involvement of the brain. Analysis of the data in such cases requires labor-intensive arterial blood sampling and analysis of metabolites. Recent description of TSPO ligands that are not sensitive to polymorphism is encouraging [26]. Other currently evaluated neuroinflammation imaging targets include the cannabinoid receptors CB1 and CB2 which together constitute the endocannabinoid system [27]. Multiple ligands targeting CB2 are being developed [28,29,30] with encouraging animal applications [29]. Targeting leukocytes and platelets changes associated with different infectious processes has also been attempted. One example is the use of single-chain antibodies that recognize the ligand-induced binding sites (LIBS) of GPIIb/IIIa in activated platelets [31]. GPIIb/IIIa becomes exposed upon activation and therefore offers the opportunity to target activated platelets [32]. By conjugating the LIBS antibody to iron oxide nanoparticles, they used it in an animal model of cerebral malaria and were able to detect abnormal vascular platelet aggregation in vivo. This approach allowed imaging of the infection before pathology could be visualized by conventional in vivo MRI [31].
As an offshoot of optical imaging , two-photon microscopy has further opened the doors to better understanding of pathogen-host interaction in the brain. Up till now a handful of studies have used this technology in the evaluation of infectious processes; however the available literature points toward a big role for this technique in better understanding the role of specific immune cells such as CD+ lymphocytes and antigen-presenting cells (APCs) in the pathophysiology of CNS infection. The potential applications are limitless. For example, in the brains of mice chronically infected with Toxoplasma gondii , two-photon microscopy showed that antigen (Ag)-specific CD8 T cells were recruited and persisted in the presence of ongoing Ag recognition. The dynamics of the interactions between T cells and the parasite could then be visualized real time: while CD8 T cells made transient contacts with granuloma-like structures containing parasites and with individual CD11b+ APCs, including some that did not contain parasites, they ignored intact Ag-bearing cysts, astrocytes, and neurons, including infected neurons [33]. In another example, two-photon imaging was used in mice infected with different strains of malaria, showing that CD8+ T cells tended to accumulate mainly in the perivascular spaces of experimental cerebral malaria (ECM) mice . This however was also seen with strains that did not cause ECM, indicating that the presence of perivascular T cells per se is insufficient to cause ECM [34]. What differentiated the strains however were dynamic time-lapse movies that showed distinct differences in T-cell motility rather than localization, with the arrest of T cells in the perivascular spaces correlating with the pathogenic potential [34]. This dynamic real-time demonstration of host–pathogen (immune system) interactions within living tissue will surely facilitate and improve our understanding of immune responses to persistent pathogens in the brain.
8.5 Summary
Imaging of infection in the CNS has been handled using cross-sectional imaging for more than two decades now resulting in a large array of descriptive diagnostic criteria, capable, in most circumstances of narrowing the differential diagnosis, detecting life-threatening complications and establishing baseline for assessment of treatment response. Limitations however exist, and in many circumstances, both cross-sectional imaging and nonspecific molecular imaging, such as 18F-FDG, fail to establish a diagnosis. The availability of pathogen-specific imaging agents/ligands would have a great effect on the management of patients with CNS infection. Besides early diagnosis, avoidance of diagnostic brain biopsies can have significant effect on the mortality and morbidity of patients.
References
Buursma, A.R., et al., [18F]FHPG positron emission tomography for detection of herpes simplex virus (HSV) in experimental HSV encephalitis. J Virol, 2005. 79(12): p. 7721-7.
Weinstein, E.A., et al., Imaging Enterobacteriaceae infection in vivo with 18F-fluorodeoxysorbitol positron emission tomography. Sci Transl Med, 2014. 6(259): p. 259ra146.
Li, Z.B., et al., The synthesis of 18F-FDS and its potential application in molecular imaging. Mol Imaging Biol, 2008. 10(2): p. 92-8.
Cook, S.H. and D.E. Griffin, Luciferase imaging of a neurotropic viral infection in intact animals. J Virol, 2003. 77(9): p. 5333-8.
Hwang, S., et al., Persistent gammaherpesvirus replication and dynamic interaction with the host in vivo. J Virol, 2008. 82(24): p. 12498-509.
Luker, G.D., et al., Noninvasive bioluminescence imaging of herpes simplex virus type 1 infection and therapy in living mice. J Virol, 2002. 76(23): p. 12149-61.
Luker, G.D., et al., In vitro and in vivo characterization of a dual-function green fluorescent protein--HSV1-thymidine kinase reporter gene driven by the human elongation factor 1 alpha promoter. Mol Imaging, 2002. 1(2): p. 65-73.
Zaitseva, M., et al., Application of bioluminescence imaging to the prediction of lethality in vaccinia virus-infected mice. J Virol, 2009. 83(20): p. 10437-47.
Luker, K.E., et al., Bioluminescence imaging of vaccinia virus: effects of interferon on viral replication and spread. Virology, 2005. 341(2): p. 284-300.
Raaben, M., et al., Non-invasive imaging of mouse hepatitis coronavirus infection reveals determinants of viral replication and spread in vivo. Cell Microbiol, 2009. 11(5): p. 825-41.
Kang, H.R., et al., Persistent infection of a gammaherpesvirus in the central nervous system. Virology, 2012. 423(1): p. 23-9.
Li, X.F., et al., Noninvasive bioluminescence imaging of dengue virus infection in the brain of A129 mice. Appl Microbiol Biotechnol, 2013. 97(10): p. 4589-96.
Phillips, A.T., et al., Bioluminescent imaging and histopathologic characterization of WEEV neuroinvasion in outbred CD-1 mice. PLoS One, 2013. 8(1): p. e53462.
D’Archivio, S., et al., Non-invasive in vivo study of the Trypanosoma vivax infectious process consolidates the brain commitment in late infections. PLoS Negl Trop Dis, 2013. 7(1): p. e1976.
Dellacasa-Lindberg, I., N. Hitziger, and A. Barragan, Localized recrudescence of Toxoplasma infections in the central nervous system of immunocompromised mice assessed by in vivo bioluminescence imaging. Microbes Infect, 2007. 9(11): p. 1291-8.
Mook-Kanamori, B.B., et al., Daptomycin in experimental murine pneumococcal meningitis. BMC Infect Dis, 2009. 9: p. 50.
Myburgh, E., et al., In vivo imaging of trypanosome-brain interactions and development of a rapid screening test for drugs against CNS stage trypanosomiasis. PLoS Negl Trop Dis, 2013. 7(8): p. e2384.
Pichler, A., et al., Generation of a highly inducible Gal4-->Fluc universal reporter mouse for in vivo bioluminescence imaging. Proc Natl Acad Sci U S A, 2008. 105(41): p. 15932-7.
Yull, F.E., et al., Bioluminescent detection of endotoxin effects on HIV-1 LTR-driven transcription in vivo. J Histochem Cytochem, 2003. 51(6): p. 741-9.
Kadurugamuwa, J.L., et al., Reduction of astrogliosis by early treatment of pneumococcal meningitis measured by simultaneous imaging, in vivo, of the pathogen and host response. Infect Immun, 2005. 73(12): p. 7836-43.
Venneti, S., et al., Longitudinal in vivo positron emission tomography imaging of infected and activated brain macrophages in a macaque model of human immunodeficiency virus encephalitis correlates with central and peripheral markers of encephalitis and areas of synaptic degeneration. Am J Pathol, 2008. 172(6): p. 1603-16.
Doorduin, J., et al., [11C]-DPA-713 and [18F]-DPA-714 as new PET tracers for TSPO: a comparison with [11C]-(R)-PK11195 in a rat model of herpes encephalitis. Mol Imaging Biol, 2009. 11(6): p. 386-98.
Fujita, M., et al., PET reveals inflammation around calcified Taenia solium granulomas with perilesional edema. PLoS One, 2013. 8(9): p. e74052.
Lee, D.E., et al., Lack of neuroinflammation in the HIV-1 transgenic rat: an [(18)F]-DPA714 PET imaging study. J Neuroinflammation, 2015. 12(1): p. 171.
Owen, D.R., et al., An 18-kDa translocator protein (TSPO) polymorphism explains differences in binding affinity of the PET radioligand PBR28. J Cereb Blood Flow Metab, 2012. 32(1): p. 1-5.
Zanotti-Fregonara, P., et al., Synthesis and evaluation of translocator 18 kDa protein (TSPO) positron emission tomography (PET) radioligands with low binding sensitivity to human single nucleotide polymorphism rs6971. ACS Chem Neurosci, 2014. 5(10): p. 963-71.
Haugh, O., et al., The emerging role of the cannabinoid receptor family in peripheral and neuro-immune interactions. Curr Drug Targets, 2016. 17: p. 1-8.
Ahmad, R., et al., Whole-body biodistribution and radiation dosimetry of the cannabinoid type 2 receptor ligand [11C]-NE40 in healthy subjects. Mol Imaging Biol, 2013. 15(4): p. 384-90.
Savonenko, A.V., et al., Cannabinoid CB2 Receptors in a Mouse Model of Abeta Amyloidosis: Immunohistochemical Analysis and Suitability as a PET Biomarker of Neuroinflammation. PLoS One, 2015. 10(6): p. e0129618.
Slavik, R., et al., Discovery of a high affinity and selective pyridine analog as a potential positron emission tomography imaging agent for cannabinoid type 2 receptor. J Med Chem, 2015. 58(10): p. 4266-77.
von Zur Muhlen, C., et al., A contrast agent recognizing activated platelets reveals murine cerebral malaria pathology undetectable by conventional MRI. J Clin Invest, 2008. 118(3): p. 1198-207.
Schwarz, M., et al., Conformation-specific blockade of the integrin GPIIb/IIIa: a novel antiplatelet strategy that selectively targets activated platelets. Circ Res, 2006. 99(1): p. 25-33.
Schaeffer, M., et al., Dynamic imaging of T cell-parasite interactions in the brains of mice chronically infected with Toxoplasma gondii. J Immunol, 2009. 182(10): p. 6379-93.
Shaw, T.N., et al., Perivascular Arrest of CD8+ T Cells Is a Signature of Experimental Cerebral Malaria. PLoS Pathog, 2015. 11(11): p. e1005210.
Author information
Authors and Affiliations
Corresponding author
Editor information
Editors and Affiliations
Rights and permissions
Copyright information
© 2017 Springer International Publishing AG
About this chapter
Cite this chapter
Hammoud, D.A. (2017). Neuroimaging. In: Jain, S. (eds) Imaging Infections . Springer, Cham. https://doi.org/10.1007/978-3-319-54592-9_8
Download citation
DOI: https://doi.org/10.1007/978-3-319-54592-9_8
Published:
Publisher Name: Springer, Cham
Print ISBN: 978-3-319-54590-5
Online ISBN: 978-3-319-54592-9
eBook Packages: MedicineMedicine (R0)