Abstract
Neural foramina stenosis (NFS), as a common spine disease, affects \(80\,\%\) of people. Clinical diagnosis by physicians’ manual segmentation is inefficient and laborious. Automated diagnosis is highly desirable but faces the class overlapping problem derived from the diverse shape and size. In this paper, a fully automated diagnosis approach is proposed for NFS. It is based on a newly proposed synchronized superpixels representation (SSR) model where a highly discriminative feature space is obtained for accurately and easily classifying neural foramina into normal and stenosed classes. To achieve it, class labels (0:normal,1:stenosed) are integrated to guide manifold alignment which correlates images from the same class, so that intra-class difference is reduced and the inter-class margin are maximized. The overall result reaches a high accuracy (\(98.52\,\%\)) in 110 mid-sagittal MR spine images collected from 110 subjects. Hence, with our approach, an efficient and accurate clinical tool is provided to greatly reduce the burden of physicians and ensure the timely treatment of NFS.
You have full access to this open access chapter, Download conference paper PDF
Similar content being viewed by others
Keywords
These keywords were added by machine and not by the authors. This process is experimental and the keywords may be updated as the learning algorithm improves.
1 Introduction
Neural foramina stenosis (NFS) is known as a common result of disc degeneration due to age. For example, about \(80\,\%\) of people suffer lower back pain caused by NFS [1, 2]. Existing clinical diagnosis by physicians’ manual segmentation is very inefficient and tedious. Automated diagnosis, which predicts class label (0:normal,1:stenosed) for a given neural foramina image, is highly desirable.
However, automated diagnosis is still challenging due to the difficulty in extracting very discriminative feature representation from extremely diverse neural foramina images [1]. This diversity leads to severe inter-class overlapping problem when classifying neural foramina images into normal or stenosed class(see Fig. 1(a)). Class overlapping problem is regarded as one of the toughest pervasive problems in classification [3–6], and severely affects the diagnosis accuracy of neural foramina. To solve it, a discriminant feature space maximizing the inter-class margin between normal and stenosed class is needed.
In this paper, a fully automated and reliable diagnosis framework is proposed for NFS. For reliable classification, it construct a new discriminative feature space (as shown in Fig. 1(d)) using a new synchronized superpixels representation (SSR) model (as shown in Fig. 1(b) and (c)). SSR model integrates class label into manifold approximation and alignment to obtain the joint decomposing, synchronizing, and clustering the spectral representation for neural foramina images from the same class. The obtained normal SSR and stenosed SSR are new superpixel representation synchronized for image from the same class. As the synchronization of SSR is merely performed for images from the same class so that images from different classes have unsynchronized superpixel representations which enlarge the inter-class difference. Hence, the constructed SSR space is highly discriminative due to the enlarged inter-class margin and reduced intra-class margin (as shown in Fig. 1(d)). With this discriminative space, any classifier, even the simple knn, could achieve superior performance in automated diagnosis of NFS. With our diagnosis framework, an automated and accurate clinical diagnosis tool is provided for NFS.
2 Spectral Graph, Spectral Bases, and Superpixels
There are three key concepts used in our framework:
Spectral graph \(\mathcal {G} = (\mathcal {V},\mathcal {E})\) is a graph structure for the pairwise similarities among all pixels within an image [7, 8]. For an image I with total N pixels, we construct \(\mathcal {G} = (\mathcal {V},\mathcal {E})\) where \(\mathcal {V}(N = |\mathcal {V}|)\) is the pixel set and each edge \(e \in \mathcal {E}\) connects two arbitrary pixels i, j in the image. Each e for i, j is weighted by W(i, j) determined by intensity, spatial location, and the contour interventions between two pixels:
where \(x_i, x_j\) are the location of the pixels i, j and the \(I_i, I_j\) are their intensities respectively. Edge(x) represents an edge detector (i.e., Canny detector) in location x. \(\delta _x, \delta _I, \delta _E\) are constants that will be assigned empirically. In practice, spectral matrix W will only be computed in k-nearest neighbors, thus W is a sparse matrix.
Spectral bases \(U = [\xi _1(\mathcal {G}),..., \xi _N(\mathcal {G})]\) are the eigenvectors of spectral matrix W [8, 9]. In practice, they are decomposed from the graph Laplacian L instead of W:
where \(Id=diag(1,1,...,1)\) is the identify matrix, D is the diagonal matrix whose elements are the row summations of W.
Superpixels are the clusters obtained from grouping images pixels based on spectral bases which approximate manifold of an image. They correspond to high level representation of an image, such as smooth and non-overlapping regions in the image.
3 Methodology
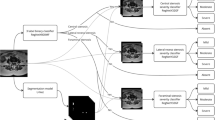
The overview of our diagnosis framework includes two phases (as shown in Fig. 2): (1) in training (Sect. 3.1), SSR space is constructed by label-supervised synchronization of spectral bases’ decomposition and clustering; (2) in testing (Sect. 3.2), the class label of an unlabeled localized neural foramina is predicted by searching its nearest neighbors in SSR space.
3.1 Training Phase
Given training set \(\{\mathcal {I},\mathcal {C}\}\) include \(M_1\) normal neural foramina images \(\mathcal {I}_{nor}=\{I_m|C_m=0, I_m\in \mathcal {I}, C_m\in \mathcal {C}, m=1,...,M_1\}\), \(M_2\) stenosed neural foramina images \(\mathcal {I}_{ste}=\{I_m|C_m=1, I_m\in \mathcal {I}, C_m\in \mathcal {C}, m=1,...,M_2\}\), and the corresponding Laplacians set \(\mathcal {L}=\{L_m, m=1,...,M\}\), where \(M=M_1+M_2\), the construction of SSR space includes the following two steps:
Spectral Bases Synchronization: Synchronized spectral bases for normal and stenosed images are simultaneously obtained by the integration of class labels \(\{C_m=C_l\}\) into Joint Laplacian Diagonalization with Fourier coupling [7]. For looking a set of synchronized bases \(\{Y_i:Y_m^TY_m=Id\}_{m=1}^M\), \(Y_m^TL_mY_m\) are approximately diagonal for \(m=1,...,M\). To ensure that the bases from the same class behave consistently, the label-supervised coupling constraints [7] are introduced: given a vector \(f^m\) on manifold of image \(I_m\), and a corresponding vector \(f^l\) on manifold of image \(I_l\), if \(C_m=C_l\), we require that their Fourier coefficients in the respective bases coincide, \(Y_mf^m=Y_lf^l\). So, the label-supervised coupled diagonalization problem can be rewritten as
where \(\varLambda _m=diag(\lambda _1,...,\lambda _K)\) denotes the diagonal matrix containing the first smallest eigenvalues of \(L_m\). F is an arbitrary feature mapping that maps a spectral map to a fixed dimension feature vector. The optimal results \(Y_1^*,...,Y_M^*\) can be classified as normal and stenosed synchronized spectral bases according to their class labels.
In practice, to resolve the ambiguity of \(Y_m\) and simplify the optimization, the first K vectors of the synchronized spectral bases are approximated as a linear combination of the first smallest \(K'\ge K\) eigenvectors of \(L_m\), denoted by \(U_m = [\xi _1(\mathcal {G}_m),..., \xi _K(\mathcal {G}_m)]\). We parameterize the synchronized spectral base \(Y_m\) as \(Y_m=U_mA_m\), where \(A_m\) is the \(K'\times K\) matrix of linear combination coefficients. From the orthogonality of \(Y_m\), it follows that \(A_m^TA_m=Id\). Plugging this subspace parametrization into Eq. (3), where \(\tilde{\varLambda _m}\) is the diagonal matrix containing the first \(K'\) eigenvalues of \(L_m\):
The solution of problem Eq. (4) can be carried out using standard constrained optimization techniques. As the label-supervised Coupled Diagonalization of Laplacians enables the approximation and alignment of manifolds for images from the same class, the obtained synchronized spectral bases approximate manifold of each image and align them for images from the same class.
Superpixels Synchronization: Normal SSR and stenosed SSR are respectively achieved by grouping all images pixels with the corresponding synchronized spectral bases. As the obtained spectral bases are automatically synchronized for images from the same class, so the obtained normal SSR and stenosed SSR simultaneously minimize the intra-class difference. Correspondingly, the unsynchronized spectral bases for images from different classes enables the obviously different superpixel representations to maximize the inter-class margin. Hence, the obtained SSR space provides a new discriminate ability for reliable diagnosis even using a simple classifier.
3.2 Testing Phase
In testing, unlabeled neural foramina \(I_w\) is first localized by a trained SVM subwindow localization classifier implemented by method introduced in [10], then its class label is predicted by finding the nearest neighbors in SSR space.
Incremental Spectral Bases Synchronization: For unlabeled neural foramina \(I_w\), incremental synchronization is proposed to obtain its mapping point \(Y_w\) in SSR space:
where \(L_w\) is the Laplacian matrix of \(I_w\), \(\{Y_m|C_m=1, m=1,...,M_2\}\) are the learned stenosed synchronized spectral bases, \(\{Y_m|C_m=0, m=1,...,M_1\}\) are the learned normal synchronized spectral bases, \(cost_{ste}\) and \(cost_{nor}\) denote the mapping cost loss. Incremental spectral bases synchronization maps \(I_w\) into SSR space.
Diagnosis: The class label of \(I_w\) is predicted by comparing the computed cost loss \(cost_{ste}\) and \(cost_{nor}\). For example, if \(cost_{nor}\) is smaller, its approximated manifold \(Y_w\) is more similar to images from normal class and the mapping point of \(I_w\) in SSR space is in normal class. Hence, the class label of \(I_w\) is naturally predicted by the minimal mapping cost loss:
4 Experiments and Results
4.1 Experiment Setup
Following the clinical standard, our experiments are tested on 110 mid-sagittal MR lumbar spine images collected from 110 subjects including healthy cases and patients with NFS. These collected MR scans are scanned using a sagittal T1 weight MRI with repetition time (TR) of 533 ms and echo time (TE) of 17 ms under a magnetic field of 1.5 T. The training sets includes two types: (1) NF images and non-NF images used to train SVM-classifier localization; (2) normal NF images and stenosed NF images used to train SSR model. These training images were manually cropped and labeled by physician according to the clinical commonly diagnosis criterion [1]. The classification accuracy, specificity, and sensitivity are reported in the average from ten runs of leave-one-subject-out cross-validation.
4.2 Results
The higher accuracy achieved by the proposed framework both in localization (\(99.27\,\%\)) and classification (\(98.52\,\%\)) are shown in Table 1. Besides, its robustness in localizing and diagnosing neural foramina with different appearance, shape, and orientation is qualitatively displayed in Fig. 3. This high accuracy and robustness is derived from the intrinsic class separation captured by our framework. Hence, an accurate and efficient diagnosis of NFS is obtained regardless of the disturbance from appearance, shape, and orientation.
Table 2 demonstrates that SSR achieved highest accuracy (>95 %) than other five classical features (<82 %) in three typical classifiers: k nearest neighbors (KNN) [5], linear discriminant analysis (LDA) [4], and support vector machine (SVM) [3]. The superiority of SSR is from its constructed discriminative feature space where images from the same class are correlated by the synchronized superpixel representation while images from different classes are separated by the unsynchronized superpixel representation. This brings a good class separation for solving the challenging class overlapping problem in automated diagnosis of NFS (see Fig. 4), which leads to the low accuracy of the other five image feature methods. In addition, such discriminative ability is so powerful that it enables the simple classifier like KNN still achieve a higher accuracy. Hence, SSR provides a reliable diagnosis framework for NFS, and can replace conventional image representation methods to be fed into other state-of-the-art classifiers for improving their accuracy and learning performance.
5 Conclusions
In this paper, we propose a novel automated diagnosis for NFS, with a new SSR model to generate discriminative feature space for reliable diagnosis. With SSR space, the class overlapping problem is overcome, and the high diagnosis accuracy of the proposed framework is achieved even using a simple KNN classifier. Hence, an efficient and reliable diagnosis tool is obtained to reduce the workload of radiologists and provide timely treatment of NFS.
References
Lee, S., Lee, J.W., Yeom, J.S., Kim, K.-J., Kim, H.-J., Chung, S.K., Kang, H.S.: A practical mri grading system for lumbar foraminal stenosis. Am. J. Roentgenol. 194(4), 1095–1098 (2010)
Rajaee, S.S., Bae, H.W., Kanim, L.E., Delamarter, R.B.: Spinal fusion in the united states: analysis of trends from 1998 to 2008. Spine 37(1), 67–76 (2012)
Guyon, I., Weston, J., Barnhill, S., Vapnik, V.: Gene selection for cancer classification using support vector machines. Mach. Learn. 46(1–3), 389–422 (2002)
Ye, J., Janardan, R., Li, Q.: Two-dimensional linear discriminant analysis. In: Advances in Neural Information Processing Systems, pp. 1569–1576 (2004)
Tan, S.: Neighbor-weighted k-nearest neighbor for unbalanced text corpus. Expert Syst. Appl. 28(4), 667–671 (2005)
Tang, J., Alelyani, S., Liu, H.: Feature selection for classification: a review. In: Data Classification: Algorithms and Applications, p. 37 (2014)
Eynard, D., Kovnatsky, A., Bronstein, M.M., Glashoff, K., Bronstein, A.M.: Multimodal manifold analysis by simultaneous diagonalization of laplacians. IEEE Trans. Pattern Anal. Mach. Intell. 37(12), 2505–2517 (2015)
Cai, Y., Ali, I., Mousumi, B., Ian, C., Shuo, L.: Unsupervised freeview groupwise m3 cardiac images segmentation using synchronized spectral networks. In: MICCAI (2015)
Wang, C., Mahadevan, S.: Manifold alignment preserving global geometry. In: IJCAI (2013)
Nguyen, M.H., Torresani, L., de la Torre, F., Rother, C.: Weakly supervised discriminative localization and classification: a joint learning process. In: 2009 IEEE 12th International Conference on Computer Vision, pp. 1925–1932. IEEE (2009)
Acknowledgment
This work was supported by the NSFC Joint Fund with Guangdong under Key Project (Grant No. U1201258).
Author information
Authors and Affiliations
Corresponding author
Editor information
Editors and Affiliations
Rights and permissions
Copyright information
© 2016 Springer International Publishing AG
About this paper
Cite this paper
He, X., Yin, Y., Sharma, M., Brahm, G., Mercado, A., Li, S. (2016). Automated Diagnosis of Neural Foraminal Stenosis Using Synchronized Superpixels Representation. In: Ourselin, S., Joskowicz, L., Sabuncu, M., Unal, G., Wells, W. (eds) Medical Image Computing and Computer-Assisted Intervention – MICCAI 2016. MICCAI 2016. Lecture Notes in Computer Science(), vol 9901. Springer, Cham. https://doi.org/10.1007/978-3-319-46723-8_39
Download citation
DOI: https://doi.org/10.1007/978-3-319-46723-8_39
Published:
Publisher Name: Springer, Cham
Print ISBN: 978-3-319-46722-1
Online ISBN: 978-3-319-46723-8
eBook Packages: Computer ScienceComputer Science (R0)