Abstract
We present a domain-agnostic system for Question Answering over multiple semi-structured and possibly linked datasets without the need of a training corpus. The system is motivated by an industry use-case where Enterprise Data needs to be combined with a large body of Open Data to fulfill information needs not satisfied by prescribed application data models. Our proposed Question Answering pipeline combines existing components with novel methods to perform, in turn, linguistic analysis of a query, named entity extraction, entity/graph search, fusion and ranking of possible answers. We evaluate QuerioDALI with two open-domain benchmarks and a biomedical one over Linked Open Data sources, and show that our system produces comparable results to systems that require training data and are domain-dependent. In addition, we analyze the current challenges and shortcomings.
You have full access to this open access chapter, Download conference paper PDF
Similar content being viewed by others
Keywords
These keywords were added by machine and not by the authors. This process is experimental and the keywords may be updated as the learning algorithm improves.
1 Introduction
With the advent of Open Data in all aspects of enterprise and government operations, valuable information is accumulating in disparate stores. Recognizing the infeasibility of a fully integrated model, Linked Data has presented itself as a paradigm for interoperability and (partial) integration. In many scenarios, such as Smart Cities [20], enterprise data can be enriched with relevant Linked Open Data (LOD) to help users find relevant entities, as well as answering more specific information needs. In this paper, we present a Question Answering (QA) system that exploits LOD and Knowledge Graphs (KGs) extracted from tabular enterprise data to answer user queries expressed in Natural Language (NL).
Traditionally, QA has focused on unstructured information (to extract answers from text fragments in documents). Notably, the IBM Watson system [1] won the Jeopardy challenge against human experts in 2011. In Watson, structured data is used just to find additional evidence, in the form of typing of answers, spatial-temporal constraints or semantic expansion for commonly appearing entity types (e.g., countries, US presidents).
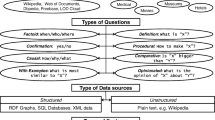
With the growth of Linked Data, various QA systems over structured semantic data have been proposed to allow end users not familiar with formal queries to express arbitrarily complex information using NL. Complementary to QA over documents, QA over large graphs is useful to find answers to factoid questions in a scenario with evolving (dynamic) semi-structured interlinked data sources, no fixed schema and no training corpora. Yet, very few systems address the fact that, often, questions can only be answered by combining information from several sources.
Driven by the QALD benchmarks [2, 3], the state of the art on QA over Linked Data has focused on answering open domain queries over the DBpedia dataset [11] and, to some extent over biomedical LOD. There are two other QA benchmarks over Freebase (which content has been recently migrated to Wikidata): WebQuestions and Free917 [17]. The Free917 benchmark includes the correct KB query and answers, while WebQuestions only provides the answers collected using crowdsourcing. These benchmarks exhibit quite different challenges: QALD involves more complex questions requiring multiple relations and operators such as comparatives and superlatives; Freebase is an order of magnitude bigger than DBpedia, and with several hundreds of possible mappings for a given keyword in a user query, a major challenge is to find the matching entities and relations in the KG. Differently from ontology-based QA over DBpedia, QA approaches over Freebase are based on IR techniques and weak-supervised learning. Often, QA systems are evaluated for only one of the benchmarks. None of the ontology-based QA approaches, many of them unsupervised, have been evaluated over both DBpedia and Freebase.
Structured QA is also gaining popularity among search engines, e.g., Google KG. With the rise of the Web of Data, more questions can be answered, thus reducing the sparseness typical of this kind of systems (for a question to be answered the knowledge need to be encoded in a KG). However, differently from industry systems that rely on a proprietary curated KG, this incremental growth comes at a cost: noise, heterogeneity, incomplete schema, and missing linkage across graphs. As a result, a large space of candidates and mapping combinations need to be checked to find translations to a user query in quasi real-time, particularly in cases where different KGs may contain only part of the answers.
In sum, the main contributions of our work are the following:
-
A practical approach outlining the most significant design choices to support practitioners in the open-domain QA. Our implemented system, QuerioDALI, combines a novel linguistic analysis built on the top of the IBM Watson NLP pipeline to obtain a set of PAS (predicate argument structures) that link the user query terms together, and a Graph Pattern (GP) search component to translate these PAS into a logical representation that coveys the meaning of the query, obtaining and ranking answers based on evidence extracted from the KGs using semantic techniques, without the need of a training corpus.
-
A novel component to merge facts (GPs) across graphs, obtained from both enterprise and Open Data. LOD is used both to extract answers or partial answers (enriching the enterprise data) and to understand the user query (through semantic expansion). It uses interlinked data to answer queries that could not be answered by a single KG.
-
We perform a benchmark-based evaluation in two scenarios to show that the system can produce comparable results to state of the art systems that require training: (a) Open domain over large and heterogeneous KGs: using both QALD-5 and Free917; (b) Based on a real-world use case in the Smarter Care domain, where relational enterprise data and biomedical LOD need to be combined to address care worker’s information needs regarding a specific client: the evaluation is based on the QALD-4 biomedical task.
In this paper, we analyze the challenges of balancing precision and recall in every step of the process: from understanding the user query to obtaining ranked interpretations leading to answers. Moreover, differences in terms of how the data is structured across graphs (e.g., Freebase vs. DBpedia) raises more challenges than scalability, regarding the mismatch between the NL query and KGs. The next section elaborates on a motivating use case. Section 3 presents the main architectural components. Section 4 presents the evaluations and open challenges, followed by related work and conclusions in Sects. 5 and 6.
2 Use Case: Smarter Care
For most enterprise applications, user interfaces are based on the premise that business analysts and designers are able to capture the information that will be required by a user and display it when requested or as part of a business process. For example, a nurse doing home calls will most likely be interested in the address of the patients, their conditions and the medication that they are taking, among others. As the body of open knowledge available expands, there is a significant opportunity to satisfy the information needs of a user when it is difficult to predict these needs. Search systems can be used to look up information, but fail when they need to combine information from an enterprise system with Open Data. For example, answering the questions What are the side-effects for all the medications of this patient? or Is any of the patient’s medication related with insomnia? requires retrieving the medications of a patient from the enterprise system, looking each one of them up individually on an online source and examining each list of side-effects.
Figure 1 shows BlueLENS, an example system to integrate multi-source information in the domain of care. The collected information is presented as a set of self-describing elements (in each rectangle) to allow care workers to obtain a 360-degree view of a client. Although beyond the scope of this paper, this system allows navigation and ranking of data from multiple sources.
But what happens when the user has an information need that is both specific and not covered by the existing configurations? How can we enrich the data that is already handled by this system with additional information about, among others, diseases, drugs and their interactions, openly available on the Web? The work in this paper has been driven by this real-world problemFootnote 1. In the following Sections, some of the examples are related to this base scenario.
3 Approach for QA Over Knowledge Graphs
We propose a QA pipeline to build up the formal graph queries needed to satisfy complex information needs, expressed in NL, from both open and enterprise KGs.
In our Smarter Care scenario, the system DALI [20] is used to lift the enterprise relational data from IBM Cúram [10] into a KG with explicit semantics and linked to well-known W3C and LOD models. We distinguish between two types of data-sources depending on how the data is interfaced by the system: (1) The QA KGs from which answers are extracted, built from open and enterprise datasets and exposed through federated SPARQL endpoints; (2) The Annotators, which are the distributed LOD sources and dictionaries used to perform query expansion. For the open-domain scenario, the QA KGs consists of DBpedia 2015-14 and Freebase. For the Smarter Care scenario it contains enterprise data from Cúram about patient conditions, prescriptions and care plans, and the biomedical ontologies SIDER [13], drugbank [8] and diseasome [7]. As Annotators, we use WordNet, the lemon lexicon (http://lemon-model.net) and schema.org for both scenarios; and DBpedia for the Smarter Care scenario. However, our system can be configured to use any ontologies or dictionaries. QuerioDALI uses a combination of off-the-shelf and novel components (see Fig. 2). In this paper we detail the following steps and components:
- Step 1: :
-
Deep NLP and Named Entity (NE) extraction. Based on the Watson parsing pipeline and off-the-shelf NE annotators to generate domain-independent PAS triples for the query.
- Step 2: :
-
Anchoring. Required to bridge the gap between the user vocabulary and the terminology in the KGs. Semantic expansion is used to find the candidate URIs.
- Step 3: :
-
Graph Pattern Search. From this large pool of candidates, a novel component translates the PAS triples into GPs, which convey into a formal query (in terms of entities, relations and logical operators) that can be executed against the KGs.
- Step 4: :
-
Merging of GPs. Merging facts and partial translations, from the same or different sources, requires finding join terms across the GPs to be joined, as well as entity co-reference if the query involves merging across graphs.
- Step 5: :
-
Ranking. Due to ambiguity, a query can have alternative representations that can be combined in different ways, based on the query type (factoid, boolean, etc.), answers are ranked using a score that reflects the confidence of the system on the evidence.
The coverage and accuracy of each component influences the design choices and performance of the next component in the pipeline.
3.1 Step 1: Deep NLP Parsing and NER
For the linguistic processing of a question, we adopted Watson’s general purpose deep parsing implemented as an Apache UIMA application. It receives as input a question and identifies syntactic, morphological and semantic elements of the question, building a dependency parse tree. The predicate arguments of a tree node are other nodes that may come from remote positions on the tree. A dependency parse tree is shown in Fig. 3.
The challenge here is generating domain-independent PAS triples in the form of <subject, predicate, object> . To do that, we use the Watson Subtree Pattern Matching framework, which allows expressing and executing rules over the dependency tree. We wrote 25 general-purpose rules to extract the triple patterns for the kind of NL sentences found in the QALD training sets for English [3]. However, its coverage can be extended. On Fig. 3 we see the PAS triples obtained for an example query and one example rule fired to detect the PAS with the superlative “lowest readmission” (pattern = nounAdjSup).
In general terms, all the rules that can be fired generate one of the following structures: (1) Basic PAS: with explicit subject-predicate-object, the subject can be a wh-term (e.g., Who climbed Mount Everest?); (2) Noun modifiers PAS for which the predicate is not explicitly given (e.g., Russian mountain); (3) PAS Tuples: those for which the predicate is associated to 3 instead of 2 modifier terms (e.g., Who climbed Mount Everest the first?); (4) PAS with a superlative/comparative determiner (e.g., highest mountain); and (5) PAS with spatial or temporal modifiers for the whole sentence (e.g., in May 2008).
NE extraction is crucial to find the syntactic roles in a sentence and multi-word terms, in the example “elderly care”. Thus, we extend the scope of Watson’s NE recognizer (based on its own domain-independent ontology) by also using LOD NE extractors to detect multi-words before parsing the sentence; in particular DBpedia Spotlight [16] and Alchemy API (www.alchemyapi.com) For each term, or NE, in the PAS we capture the following features: its syntactic role (subj, obj, pred, nadj, etc.), lemma, covered text, if it is the focus of the sentence, the Part-Of-Speech and associated features (plural/singular, verb tense, etc.). If the entity is recognized, the semantic type is also provided (city, sports team, etc.).
3.2 Step 2: Anchoring and Semantic Expansion
The PAS Triples obtained in Step 1 represent how the terms in a sentence are linked together, but it does not necessarily correspond to the way information is structured in the KGs, e.g., the linguistic predicate designed in Who designed the giant dipper? may translate to an ontological type designer, instead of (or in addition to) an ontological property. Thus, before we can translate each PAS triple into one or more GPs (from the same or different KGs), Step 2 returns all candidate entities for each term in the PAS, together with their type and a matching score, without forcing any heuristic on the kind of expected type.
The result is a Mapping Table per linguistic term, containing the candidate URIs for each underlying graph to be queried. The matching is based on exact and approximate index searches over the entity labels (rdfs:label) for instances, properties and concepts, as well as literal values. Wh-words in the PAS Triples are replaced with semantic equivalent term(s), if appropriate: person/organization for who, date for when, place/location for where.
Semantic expansion of query terms is performed using the lexicons and KGs selected for the pool of annotators, such as synonyms and hypernyms from WordNet, and other lexically related words obtained following properties such as owl:sameAs, SKOS:broader, dcterms:subject and redirects if using DBpedia. For example, the term penicillin G (in the question What are the side effects of penicillin G?) is annotated with the DBpedia term Benzylpenicillin, which has an owl:sameAs link to the entity in SIDER labeled Penicillin G Potassium Injection that links to the list of side effects that answers the question.
Lessons Learned. The entity matching problem is hard. For example, there are more than 200 entities with the exact label Founders in Freebase. Furthermore, noun-modifier PAS, composed of two terms with an implicit relation, may often refer to a compound term (multi-word) missed by the previous NE recognition. Thus, besides looking for URIs matching each independent term it also identifies if there are candidate matches for the compound (e.g., the PAS <order, ?, dragon> in Who founded the order of the dragon). While expanding the search space is needed to find the correct translation, the more accurate the matches are, the less computational effort is required to find the GPs in the next step. Thus, techniques are required to prune the space of candidate solutions:
-
A syntactic score (based on the index search and string distance metrics) is used to rank and select only those matches over a minimum threshold and up to a maximum number (that can be adjusted for each KG). In addition, unconnected properties, classes with no instances, and instances with no type are removed.
-
Approximate (fuzzy) matches are returned only if exact matches are not found for the term, including the lemma and plural forms. The score of lexically related matches (synonyms, hypernyms) is penalized with respect to matches to the original query term, as they can also introduce noise. Expansion is based on linguistic features (POS, Watson types), e.g., derived words from WordNet (e.g., Russia in Russian mountains…) are not obtained for entities which semantic type is person, such as musician (derived word: music), hypernyms are excluded in the case of proper nouns, and meronyms are only used for geographical entities.
This is the only component that requires tuning the exact and index searches according to the source to be queried. While tuning was not needed to query the biomedical ontologies, Freebase/DBpedia have some unique ways to structure data that should be considered not to miss relevant mappings: aliases (alternative labels) are presented through non-standard RDF properties. Ambiguous terms (with different URIs) in DBpedia are represented with different labels, e.g.: db:Sunflowers_(Van_Gogh_series), as well as redirects (from db:Sunflower to db:Helianthus), while in Freebase they share the same label. Properties in DBpedia may be covered by the ontology (e.g., dbo:founder), while in Freebase they have different namespaces for different domains (fb:organization.founders, fb:formula1.founders, and so on). Finally, DBpedia types are structured in a taxonomy (requiring inference through the subsumption hierarchy), while, in Freebase, instances belong to a flat list of types (where the most relevant one is given by fb:common.topic.notable.type).
3.3 Step 3: Template-Based Graph Pattern Search
The output of this component is a set of Graph-Views, one per graph. Each Graph-View consists in a set of ranked GPs. We define a GP as: (i) a set of triple patterns or BGPs (Basic Graph Patterns); (ii) JOINS among them; (iii) FILTER and modifiers such as ORDER BY, COUNT, OFFSET, LIMIT (as a query can be translated into alternative GPs, OPTIONALS are covered); (iv) the variables that are the focus (the variables we seek an answer for) of the GP; and (v) a confidence score, explained in Sect. 3.5.
GPs can be readily translated to SPARQL queries. In this step, each GP is formed with BGPs that belong to the same graph and that join together provide a translation to one or more given PAS Triples in the query. The algorithm is iterative, looking for the best translations first, and expanding the search only if required:
- Step 3.1. :
-
For each PAS Triple, KGs are sorted based on their coverage. Thus, the graphs that have candidate matches for all or most of the terms in the PAS are selected first.
- Step 3.2. :
-
For each PAS Triple and each covering graph with candidate matches, the search for GPs is performed using parametrized pattern templates. Only GPs that produce bindings after executing the associate SPARQL query are selected.
- Step 3.3. :
-
For queries with multiple PAS Triples, Graph-Views are created by merging the GPs belonging to the same graph but covering different PAS. Two GPs are merged if they have a common join (focus term) and their joining produces a non-empty set of bindings. A merged GP will have a higher ranking than a GP that could not be merged with any other, as it provides a more complete translation (coverage) to the user query.
If the Graph-View combines GPs that translate each PAS in the query (or if there is only one PAS in the query) the query can be answered within one graph and this step already generates a complete translation. If the Graph-View contains alternative translations (i.e., different interpretations of the query), answers are ranked (Sect. 3.5). However, for some queries, Graph-Views contains only partial translations and a complete translation (answers) can only be obtained by combining GPs across graphs (Sect. 3.4).
Parametrized Pattern Templates.
To execute the right pattern templates to search for the GPs providing a more meaningful representation of each PAS, while reducing the amount of queries, we use the types of the candidate matches. There are 11 direct patterns and 7 indirect patterns (corresponding to more costly SPARQL queries that are executed only if no GPs are found using the direct ones). In Table 1, we give some examples of patterns executed for a given combination of candidate typesFootnote 2.
Often, properties are either implicit or difficult to match. If there are matches not only for the subject and/or object of the PAS but also for the properties, a FILTER is added to the patterns above. If the GP does not produce any binding, the query is executed again without the filter: FILTER (?prop = <property1> || ?prop = <property2> || etc.). Schema information such as domain and range is often missing so properties need to be inferred by looking at the instance-level when invalid ontological matches are found for the property. String metrics and annotators are used to calculate semantic relatedness between all possible properties for the matched entity(-ies) and the property in the query. In particular, we use WordNet taxonomy to measure their semantic distance. Thus, the most related ontological properties to the user query term are those over a given threshold when applying Wu and Palmer formula [21] (Formula 1) and the rest are discarded. For example, in Which mountains are higher than Anapourna? (pattern 7: ?s a db:Mountain. ?s db:elevation ?o1. db:Anapourna db:elevation ?o2 FILTER (?o1 > o2)), none of the properties for the instance Anapourna could be lexically matched; the right ontological property elevation is found among all of them due to its semantic relatedness to the query term high:
Formula 1.
Relatedness between term C1 and C2, where C is the lowest common ancestor between C1 and C2, Ni is the length of the path from Ci to C, and N is the length of the path from C to the root.
Merging patterns.
These patterns also determine the way BGPs are combined to create a merged GP for the same graph. For instance, for two type-instance GPs (pattern 2) the join answers are given by the common focus: ?s, as in Which Russian rivers flow into the Black Sea? (join term: the subject river) the merged GP joins all BGPS in both GPs:
GP1: ?s rdf:type dbo:river.?s ?p1 db:Russia. FILTER (?p1 = db:sourceCountry)
GP2: ?s rdf:type dbo:river. ?s ?p2 db:BlackSea. FILTER (?p2 = db:mouth)

3.4 Step 4: Merging Across Graphs
Partial answers from GPs across graphs that are translations to different PAS Triples need to be combined to generate complete answers. For GPs to be merged they need to have at least one variable (binding) in common, as well as one BGP with a common mapping (the subject or object) that corresponds to the Join Term. However, when merging across graphs, the Join Term may not necessarily be represented by the same URI across graphs, even if they refer to the same real world entity; the same applies to the bindings in common. Therefore, merging across federated graphs presents two challenges:
-
(1)
Finding the join term between each pair of GPs to be merged considering the query.
-
(2)
On-the-fly entity co-reference based on both a syntactic (similar labels) and a semantic (semantic equivalent link) merging between the join term and bindings across graphs.
Consider an example query in the biomedical scenario: Is Eplerenone having side effects on Teresa’s conditions?, finding an answer requires combining a GP from the patients’ graph representing Teresa’s conditions and a GP from the SIDER ontology with the side effects for the drug Eplerenone. The first step is to find the join term for the query, which in this case is represented by different URIs and query terms in each graph. From the three PAS Triples: <Eplerenone, have, side effects> <side effects, conditions> <conditions, Teresa>, the first one is translated in SIDER and the third in the patient’s graph. The second PAS, without any valid translation, links Side Effects (in SIDER) and Conditions (in the patients’ graph); that is, the two terms that need to be joined to retrieve the answer.
The second step is to retrieve the common bindings for the join terms across graphs, which will likely have different URIs even if they represent the same real-world entity. We perform two types of merging: syntactic and semantic. The semantic merging relies on the linkage across entities through equivalence relations. To find the linkage between the join variables (renamed as ?s for instances of Conditions and ?sjoin for instances of Side Effects), while avoiding pairwise comparison of entities binding each join, the following BGPs are added to the merged ones (GP1 + GP2) before executing the final SPARQL query to retrieve the answers over the federated graphs:
GP1: ?s rdf:type <Condition> . <Teresa> ?p3 ?ent. ?ent p2 ?s.
GP2: ?sjoin rdf:type <Side_effects> . <drug:Eplerenone> ?p1 ?sjoin.
{?s ?rel1 ?sjoin}. UNION {?sjoin ?rel1 ?s} UNION {?sjoin ?rel1 ?same. ?s ?rel2 ?same.}
FILTER ((?rel1 == owl.sameAs || skos.closeMatch) && (?rel2 == owl.sameAs || skos.closeMatch))
The answer to the query is true, as there is an evidence graph, shown in Fig. 4, where Teresa’s Condition, prediabetes, links to one of the side effects of Eplerenone, through the DBpedia instance Diabetes_mellitus.
However, as equivalence links across entities are often sparse, we also perform syntactic merging. An index search is added to the merged GP to find those instances from each join with similar labels.
3.5 Step 5: Ranking of Answers
The answers are the bindings of the variables representing the focus of the query over a given graph. Ranking is crucial for obtaining the answers with the best confidence among all the alternative GP representations and their combination (in the same or across graphs).
Two different scores reflect how well a GP matches a PAS in a user query: (1) At BGP level: the average score of each individual mapping in a BGP is computed. A higher score is assigned to BGPs that combine more candidate matches (e.g., if besides subject/object, the property is also match); and (2) At GP level: this is based on the pattern executed – direct patterns carry more weight than indirect ones. The rationale behind it is that the longer the distance between two candidate terms, the more likely the translation is noisy. Active and passive forms are also considered, if the same relation is valid in the two directions (from subject to object and vice-versa), e.g., who was J.F Kennedy successor of?, a higher score is given to the GP with the right directionality for the relationship.
In short, as show in Formula 2, the final confidence score of each answer A, denoted by CF(A), is a combination of the score of the GPs where the answer is computed and the scores of the BGPs in such GPs, irrespective from the graphs that they are computed from:
Formula 2.
Specifically, let A be an answer in GPA and BGPA, where there are in total m GPs across all data graphs and n BGPs in GPA
Note that answers can be represented by an entity (or a list), a datatype literal, a boolean a count (how many), or blank nodes (e.g., What is the revenue of Apple? is answered in Freebase by a set of answer nodes, where each one contains a set of property-values representing the amount, currency and date). For each answer (or set of), supporting evidence can be retrieved as per user request. An evidence graph is the subset of the graph containing all the bindings and mappings the query is translated into. When partial answers are obtained across graphs, co-reference across entities is performed; therefore, an answer may correspond to more than one URI from different graphs.
4 Evaluation
We evaluate the system with a set of blind questions based on two scenarios and for each question q, we compute precision (P), recall (R) and F-measure (F1), as defined in QALD:
To evaluate the ranking when there are multiple translations, we calculate: P/R@1 by considering only the answers which confidence score is the highest, ranked in position 1; P/R@2 for the results in the first and second positions; and P/R@3 for the results from the first to the third position. The last (P/R@3), essentially considers all answers (till position 3), without ranking. Detailed results are documented online (https://goo.gl/0o0KYy).
4.1 Scenario 1: Open Domain
DBpedia QALD-5 Evaluation: We used the 2015 QALD-5 test set [3], which consists of 50 questions annotated with the corresponding SPARQL query and answers. From those 50 questions, question Q21 is missing and question Q42 is classified as out of scope for DBpedia KB. Therefore, we measure the average P, R and F1 over 48 English queries of different complexity; QALD queries may require operations beyond triple matching such as counting or ordering. Retrieving all possible answers to a given query (total recall) over large and heterogeneous sources such as DBpedia is a challenge, even for manually created gold standards. This is due to the presence of duplicated entities (same real world entity represented with different URIs), heterogeneous properties (dateOfbirth, birthdate, etc.), and literal values that should correspond to entities, but were not mapped to. QALD queries contain UNIONs to get all answers when a question can be translated into alternative valid SPARQL queries. However, for 6 of the benchmark queries, QuerioDALI found different translations leading to an extended set of valid answers. For those queries, we have updated the set of answers in the benchmark and updated the P/R accordingly (the updated queries are documented online). Take Q4: Which animals are critically endangered? the SPARQL query in QALD retrieves a total count of 1613 distinct animals as answers: Select ?uri where {?uri rdf:type db:Animal. ?uri db:conservationStatus ‘CR’ }
However, there are 1629 answers if one considers the UNION with other valid translations: {?uri rdf:type dbo:Animal . ?uri dbp:status res:Critically_endangered} UNION
{?uri dcterms:subject dbc:Critically_endangered_animals . ?uri dbo:kingdom res:Animal}
In Table 2 we present our P/R and F1 results. The set of results obtained in the first position are more precise. The best F1 of 0.61 is for the answers ranked first (F1@1), proving that our ranking mechanism ranks the best answers first. From 48 questions, the system was unable to find answers for 10 of the queries (F1 strictly 0). Only for 2 of the queries the answers were not ranked in the first position but the second. In the first position, 25 queries were answered with an F1 of strictly 1, while for 11 queries either some answers were missing (4 of them with P = 1, R = (0,1)) or inaccurate answers were retrieved (6 of them with R = 1, P = (0,1)), or both (1 of them with P = R = (0,1)). For the second and third position, recall is increased but at a cost, as more inaccurate answers are also retrieved.
QALD campaigns are notably challenging, an F1 of 0.61, even if far from perfect, is a promising result. To put it into perspective, the average F1 of QALD-4 was 0.34 while the average F1 of QALD-5 is 0.43. The relatively low values show that the complexity of the questions is still high. Note that our P, R and F1 above are with respect to the total number of questions. This is what QALD reports as global-measures, which considers all the questions and not just the processed questions (those for which a system is able to provide a query or an answer, even if wrong). Our system is designed to give results even if only partial results are found (e.g., in Q38 Where did the architect of the Eiffel Tower study? QuerioDALI finds who is the architect of the Eiffel Tower but not where he studied, giving partial results rather than no answers). Thus, QuerioDALI only fails to process 3 out of the 48 questions, reaching an F1 - if only processed questions are considered- of 0.84.
Xser [12] has the best global F1 reported in QALD-5: 0.63, just 0.02 over our F1@1. This is well over the second ranked system with a global F1 of 0.3. Like our system, Xser analyzes the query through a semantic parser, and then instantiates the query with respect to the KB. However, differently from ours, Xser requires training data as it relies on a structure prediction approach implemented using a Collins-style hidden perceptron.
Freebase Free917 Evaluation: We used the first 101 queries from Free917 to evaluate and manually analyse why some queries failed to reach P/R of 1; 14 of those queries did not produce any answer (out of scope using the last available Freebase data dump). Thus, we measure the average P, R and F1 over the remaining 87 queries covering a wide range of domains. We did not use any manually crafted lexicons tailored to answer these queries [9], as addressing entity recognition and anchoring without domain adaptation is an integral part of the challenge we want to evaluate.
As shown in Table 2, the best F1 of 0.72 is for the answers ranked first (F1@1). The results obtained in the first position are more precise, with a small drop on recall compared with position 2. There is not significant drop on precision between position 2 and 3. Interestingly this is because fewer alternative translations are generated when using Freebase instead of DBpedia. We believe F1 is better for this benchmark because Free917 queries tend to be more tailored to the underlying KG than QALD queries. However, while they are linguistically less complex (based on a single relation without comparisons or superlatives) they can generate complex GPs due to the presence of intermediate (blank) nodes.
The system was unable to find correct answers for 22 out of 87 queries (F1 strictly 0). In the first position, 60 queries were answered with an F1 of strictly 1, while for 1 query some answers were missing (P = 1, R = (0,1)) and 3 queries retrieved inaccurate answers (R = 1, P = (0,1)). Only for 1 query the answers were not ranked in the first position but the second, thus R@2 slightly increases by 0.006, but with a drop in P of 0.08.
OPEN CHALLENGES: Entity identification is a crucial step for QA over KGs. Existent tools to extract NEs while very useful are not enough to retrieve all relevant entities to translate a user query. We reflect on this challenge with a small experiment to compare the P/R of the URIs retrieved by just using DBpedia Spotlight and QuerioDALI’s final selection of URIs to semantically interpret the user questions, using as a ground truth the entities in the QALD-5 SPARQL queries. We witnessed a significant jump in P/R from QuerioDALI results over just using Spotlight: the average P was improved from 0.31 to 0.61 and the average R from 0.44 to 0.71. Note that QuerioDALI uses Spotlight in Step 1, thus this shows the baseline over which QuerioDALI had to improve to answer the user questions, i.e., by considering the semantic linkage among query terms.
Next, we analyze the reasons behind the failed queries and those with inaccurate or missing answers (F1@1 < 1). Errors may be introduced by the different components in the pipeline, in each step of the process. Thus, a query may fail for more than a reason:
Linguistic coverage.
When the system fails to linguistically understand the query and correctly represent how the terms link together. This occurs if the dependency parse tree generated for the query is incorrect or if the generic rules used to create the PAS triples from the tree do not cover a particular type of sentence. Only one DBpedia query, db40: What is the height difference between Mount Everest and K2? and one ill-formed Freebase query (fb17:How many beers come a can?) failed because the rules could not capture the linguistic dependencies. These kind of errors need to be solved by extending the coverage.
NER + Anchoring.
When the system fails to bridge the lexical gap between the NL expressions and the data: a total of 13 queries in DBpedia and 24 in Freebase. In some cases the anchoring can recover if a multi-word NE is not captured by the annotators – for fb27:When was home depot founded? it finds the entity db:date_founded combining the property and the subject - but 7 out of 24 Freebase errors are due to NEs not found. In the worst case, missing the right anchoring leads to no answers or wrong answers (db:4/13, fb:21/24), such for db8:Is Barack Obama a democrat? where the term democrat could not be mapped to db:Democratic_Party_(United_States) and it was mapped instead to the hypernym db:politician, therefore the true answer is based on not accurate enough evidence (P/R = 0). Inaccurate anchoring often leads to a combination of noisy answers among good answers (db:7/13, fb:2/24), or a loss in recall because relevant properties or instances were not mapped (db:2/13, fb:1/24). For example, in db11:Who killed John Lennon?, QuerioDALI can not find the property (db:convinction) and it returns all the instances of a person related to the two instance matches db:John_Lennon and db:Death_of_John_Lennon (only the latter is related to John Lennon’s killer), obtaining a R@1 of 1 but a P@1 of 0.09.
Synonym features are important for both datasets. WordNet semantic relatedness is useful to find the right properties, such as weigh for heavy in db48:Who is the heaviest player of the Chicago Bulls?. Hypernyms are also needed to map players to db:person and find the relevant property for heaviest (the exact mapping for player does not lead to any valid GPs). However, the use of approximate (index) matches, aliases and alternative names (e.g., foaf:name) tends to be noisy. Thus, we only use them if non exact matches are found for a term. The use of manually or semi-automatically created lexicons (if training data is available) or through user feedback, could alleviate the anchoring problem.
Template coverage.
When the system fails to bridge the structural gap between the NL expressions and the data. Queries fail if there is not a template to properly translate the query and bridge the structural gap, 5 queries failed in DBpedia due to this (0 in Freebase), of which 4 of them involved temporal reasoning in one way or another. For example, queries involving the temporal adjectives, such as youngest/oldest, that were not mapped to the relevant property db:birth-date can be partially answered by searching among the properties with datatype date, but it also obtains unrelated properties like db:death-date; Queries with since, such as since 2000, require to add a filtering pattern to find dates whose year is over 2000 (FILTER (year(?date) > = 2000)). The query db49:Show me everyone who was born on Halloween is particularly challenging as there is not temporal adjective such as “same date/year as” to indicate we are looking for persons born on the same date as they day in which Halloween is celebrated (as in the correctly answered query db14:Which artists were born on the same date as Rachel Stevens?)
Merging.
In this scenario, merging across sources is not required, even if DBpedia uses different vocabularies, they are all interlinked as part of the same graph. However, queries with more than one PAS Triple require that the respective GPs are combined in order to obtain a complete answer. In this evaluation 18 DBpedia queries require to fuse GPs, where only one query failed (db22): the system found the right GPs but the relevant property contained a String literal, with comma separated values, that could not be linked.
Ranking.
There were only 2 ranking errors in Freebase, mostly for ambiguous queries such as fb22:Who designed the Parthenon?, where the two translations retrieve the architect for two different instances of Parthenon - the right and expected one, the Parthenon in Athens, and a full scale replica in Tennessee. For queries evaluated in a federated manner, the quality can be improved by giving a higher rank when the union of answers from each data graph forms the final answers (i.e., most popular answers across graphs).
4.2 Scenario 2: Smarter Care
We use the QALD-4 Task 2 over biomedical data, based on the ontologies SIDER, Drugbank and Diseasome. There is no benchmark to evaluate patient-based questions that can be answered using our enterprise dataset and Open Data, but, nonetheless, QALD-4 Task2 includes questions that need to be answered by combining facts from graphs, thus, it allows us to evaluate the merging. We used the training dataset used to evaluate the SINA system [18], which also provides QA over interlinked datasets and high P/R measures, using a trained Hidden Markov Model for the domain. For one of the benchmark queries (Q5), we were able to find different translations due to duplicated entities leading to an extended set of valid answers, not present in QALD-4. The results are given in Table 3.
As expected, the results on a domain-specific set up are better than for open-domain QA, with an F1 of 0.85 (the SINA system obtains an F1 of 0.89). From the 25 queries in this task, 7 of them can only be answered by merging GPs across two graphs. Only for one of them QuerioDALI could not get an F1 of 1 (Q19) but, as analyzed below, the reason was not because of the merging. The system can find answers for 23 out of 25 queries (92 % coverage). The number of queries with F1 of strictly 1 are 21, thus for 84 % of the queries, perfect answers are retrieved in the first position. For two queries, the system was not able to retrieve any answer. The rest of the queries introduce noisy answers together with the good answers. We analyze the reasons behind the inaccurate results:
Linguistic coverage.
For one query, Q19: Which are the drugs whose side effects are associated with the gene TRPM6?, the linguistic component failed to retrieve one of the PAS needed to fully understand the query. From the two PAS retrieved <drugs, associate, gene RTPM6> <side effects, associate, gene RTPM6> only the second is correct. The system can recover from ambiguous associations by using the KGs to find the right linkage, but it can hardly recover if an association is missing, in this case <drugs, side effects> .
NER + Anchoring.
In domain specific scenarios, ambiguity and errors due to anchoring are notably reduced, as long as the system can rely on annotator sources with a good coverage of the domain vocabulary, in order to find both the right lexically related words and NEs (multi-words). In particular, Q14: Give me drug references of drugs targeting Prothrombin failed, because the system was unable to detect the multi-word drug reference. The system could retrieve all drugs targeting Prothrombin, but failed to find the references (F1 = 0). In Q17: Which are possible drugs against rickets? the valid answers are ranked in the second position, this is because the relation against was not mapped to any ontological property and the system ranks first all drugs with side effect on rickets (from SIDER) and after all drugs with diseasome:possibleDrugs to rickets (the valid answers).
Templates coverage.
Bridging the structural gap is particularly challenging when schema information is missing. This is the case for Q10:Which foods does allopurinol interact with? (F1@1 = 0.35), where the interact maps to the ontological properties for allopurinol: drugbank:interactionDrug and drugbank:foodInteraction. The system retrieves all the values as answers, but only the latter leads to precise answers. This is because the relevant property is a datatype and the system can not infer based on the type food.
5 Related Work
QA approaches over Linked Data are popular because they balance expressivity with posing queries in an intuitive way, using NL. Here we look at approaches that are agnostic to the underlying KG or can scale to large open domain scenarios.
On Generating Templates, user questions are generally mapped into appropriate artifacts that can be processed over RDF datasets. One type of mapping is from questions to triples, e.g., PowerAqua [19], and FREyA [5]. FreyA leverages users’ feedback for disambiguating the right relation and improve accuracy over time, while PowerAqua ranks across alternative representations. However, complex questions that require performing aggregations or comparisons are not mapped in these systems. An alternative mapping is from questions to SPARQL query templates to capture more complex structures. For instance, TBSL [4] uses 22 manually curated templates to match a parsed user question into a SPARQL template, which is then instantiated using the domain vocabulary. However, capturing the semantics of input questions can lead to too rigid templates to fix the triple structure, queries with unknown syntactic constructions or domain-independent expressions cannot be parsed (F1 of 0.62 on 39 questions that could be processed out of the 50 in QALD-1). In Yahya et al. [14], phrases in the questions are extracted and matched to RDF entities using a large dictionary. To optimize the way of yielding SPARQL queries, an integer linear program has been used. The SINA system [18] parses questions into a list of keywords and links keywords to resources across datasets to form triple patterns using a Hidden Markov Model. Differently, our approach does not require training (as in [18]) or building indexes over all relation phrases in a paraphrase dictionary (as in [14]).
On Merging and ranking, only a few existing systems consolidate candidate answers among a collection of inter-connected, distributed datasets, considering the fact that sometimes answers to a question can only be provided if information from several sources are combined. Specifically, PowerAqua can query distributed sources without requiring federation, but it imposes an overhead for merging the data by performing entity co-reference (linear to the number of answers). The SINA system constructs federated SPARQL queries leveraging the built-in owl:sameAs property as linkage across entities.
On Entity Recognition against Freebase, the systems mainly use IR techniques to score answers. The Aqqu system [9] performs entity identification, template generation, relation matching and ranking. It reports an F 1 of 0.65 using Free917 without lexicon and 0.76 with a manually crafted lexicon (from which 276 questions are tested and 641 used as training). Supervised learning is used to find the mappings. Assuming all mappings are found, they only required the use of 3 templates to map the candidate instances and relations to queries. YodaQA [15] uses both Freebase and DBpedia, however, it has been designed to answer questions over text (filtering the passages containing the most clues from all NEs and noun phrases), which is why it only reaches a F 1 of 0.18 in QALD-5 [3].
6 Conclusions
Our system performs at the same level as the state of the art systems without the use of training data, both for open scenarios on the Web of Data or domain specific, particularly in the biomedical domain, where a large number of specialized ontologies are available [6].
Large scale deployment to scenarios with large evolving data and graphs, requires (besides supporting concurrency) strategies for iteratively pruning the large search space of solutions (and speed up the search for GPs by considering fewer candidates), while keeping the most promising ones based on the context of the query. In turn, (1) NL queries are converted into PAS Triples to focus on finding the entities and links that matter; relevant graphs are selected based on their coverage (not for the whole query but for a given PAS); (2) unconnected and less promising mappings are filtered out based on their syntactic relevancy (exact vs. approximate); (3) remaining mappings are assigned a confidence score based on their semantic relation to the query term (synonyms, etc.); (4) templates are then used to semantically validate the mappings according to their type, and search first for the GPs that more accurately represent the user query, extending the coverage to indirect relations only if nothing is found; (5) GPs are merged according to the entities to be joined, merging across graphs is based on semantic linkage or label similarity.
As shown in the evaluation, a major challenge is the anchoring of query terms to the ontological terms that would lead to the answer. Inevitably, the use of fuzzy search and semantic expansion to bridge the lexical gap also introduces lots of irrelevant mappings that, in some cases, may lead to GPs with inaccurate translations. Thus, ranking is crucial to compare across the possible solutions and select the translations with higher accuracy and better coverage for the query. The second challenge for both scenarios is coverage: we can incrementally add new patterns to increase the complexity of the questions, accommodating more complex queries requiring temporal reasoning, some basic statistical analysis or negations. However, covering the whole spectrum of different manners in which users express a question is an open problem. While QuerioDALI reaches F-measures comparable to systems that use training data, we believe that for these systems to overcome the lexical-structural gap and reach higher P/R over novel questions, they require a certain level of domain training. As the availability of training data is sometimes challenging, especially when dealing with sensitive information, we believe that the system should evolve into a cognitive system, with the ability to learn over time. This can be done if the system leverages users’ feedback, using a hybrid approach between open factoid QA and guided explorative queries. In turn, the learning will not only improve the QA but it can also be used to augment the KGs with relevant connections.
Notes
- 1.
BlueLENS: https://ibm.biz/Bd465t and QuerioDALi video demos at: https://ibm.biz/BdHEwF.
- 2.
Due to space constraints a full list of templates is presented in https://ibm.biz/BdHEwF.
References
Kalyanpur, A., Boguraev, B., Patwardhan, S., Murdock, J.W., et al.: Structured data and inference in DeepQA. IBM J. Res. Dev. 56(3), 10 (2012)
Unger, C., Forascu, C., Lopez, V., Ngomo, A.N., Cabrio, E., Cimiano, P., Walter, S.: Question answering over linked data (QALD-4). In: Working Notes for CLEF 2014 (2014)
Unger, C., Forascu, C., Lopez, V., Ngomo, A.N., Cabrio, E., Cimiano, P., Walter, S.: Question answering over linked data (QALD-5). In: Working Notes of CLEF 2015 (2015)
Unger, C., Buhmann, L., Lehmann, J., Ngonga Ngomo, A.-C., Gerber, D., Cimiano, P.: Template-based question answering over rdf data. In: World Wide Web, WWW 2012 (2012)
Damljanovic, D., Agatonovic, M., et al.: Improving habitability of natural language interfaces for querying ontologies with feedback and clarification dialogues. Web Semant. 19(1), 21 (2013)
Weissenborn, D., Tsatsaronis, G., Schroeder, M.: Answering factoid questions in the biomedical domain. In: Workshop on Bio-Medical Semantic Indexing and QA, CLEF 2013 (2013)
DISEASOME. http://diseasome.kobic.re.kr/. Accessed April 2016
Wishart, D.S., Knox, C., Guo, A.C, Shrivastava, S., et al.: DrugBank: a comprehensive resource for in silico drug discovery and exploration. Nucleic Acids Res. 34, D668–D672 (2006)
Bast, H., Haussmann, E.: More Accurate Question Answering on Freebase. In: CIKM 2015 (2015)
IBM Cúram. http://www-03.ibm.com/software/products/en/social-programs
Lehmann, J., Isele, R., Jakob, M., Jentzsch, A., et al.: DBpedia - a large-scale, multilingual knowledge base extracted from wikipedia. Semant. Web J. 6(2), 167–195 (2015)
Xu, K., Feng, Y., Zhao, D.: Answering natural language questions via phrasal semantic parsing. In: CLEF 2014 Working Notes Papers (2014)
Kuhn, M., Letunic, I., Jensen, L.J., Bork, P.: The SIDER database of drugs and side effects. Nucleic Acids Res. 44, 1075–1079 (2016)
Yahya, M., Berberich, K., Elbassuoni, S., Weikum, G.: Robust question answering over the web of linked data. In: CIKM 2013 (2013)
Baudis, P., Sedivy, J.: Modeling of the QA task in the YodaQA system. In: CLEF 2015 (2015)
Mendes, P.N., Jakob, M., Garcıa-Silva, A., Bizer, C.: Dbpedia spotlight: Shedding light on the web of documents. In: I-Semantics (2011)
Cai, Q., Yates, A.: Large-scale semantic parsing via schema matching and lexicon extensions. In: ACL, pp. 423–433 (2013)
Shekarpour, S., Auer, S., Ngonga Ngomo, A.C., Auer, S.: SINA: Semantic interpretation of user queries for question answering on interlinked data. J. Web Semant. 30, 39–51 (2015)
Lopez, V., Nikolov, A., Fernandez, M., Sabou, M., Uren, V., Motta, E.: Merging and ranking answers in the semantic web. In: The Asian Semantic Web Conference, ASWC 2009 (2009)
Lopez, V., Stephenson, M., Kotoulas, S., Tommasi, P.: Data access linking and integration with DALI: building a safety net for an ocean of city data. In: Arenas, Marcelo, et al. (eds.) ISWC 2015. LNCS, vol. 9367, pp. 186–202. Springer, Heidelberg (2015). doi:10.1007/978-3-319-25010-6_11
Wu, Z., Palmer, M.: Verb semantics and lexical selection. In: ACL (1994)
Author information
Authors and Affiliations
Corresponding author
Editor information
Editors and Affiliations
Rights and permissions
Copyright information
© 2016 Springer International Publishing AG
About this paper
Cite this paper
Lopez, V., Tommasi, P., Kotoulas, S., Wu, J. (2016). QuerioDALI: Question Answering Over Dynamic and Linked Knowledge Graphs. In: Groth, P., et al. The Semantic Web – ISWC 2016. ISWC 2016. Lecture Notes in Computer Science(), vol 9982. Springer, Cham. https://doi.org/10.1007/978-3-319-46547-0_32
Download citation
DOI: https://doi.org/10.1007/978-3-319-46547-0_32
Published:
Publisher Name: Springer, Cham
Print ISBN: 978-3-319-46546-3
Online ISBN: 978-3-319-46547-0
eBook Packages: Computer ScienceComputer Science (R0)