Abstract
Advances in DNA sequencing (i.e., next-generation sequencing, NGS) have greatly increased the power and efficiency of detecting rare mutations in large mutant populations. Targeting Induced Local Lesions in Genomes (TILLING) is a reverse genetics approach for identifying gene mutations resulting from chemical mutagenesis. In traditional TILLING, mutation discovery is accomplished through mismatch cleavage of mutant and wild-type DNA heteroduplexes using endonucleases. This is followed by Sanger sequencing to determine the specific sequence changes. TILLING by sequencing (TBS) uses NGS to facilitate the concurrent detection and sequence characterization of mutations, which allows researchers to prioritize mutants for further analyses. NGS increases the sensitivity of mutation detection and thus improves screening efficiency by allowing the pooling of more DNAs. Here we describe a protocol for TBS using rice as an example. First, DNA from a mutant population is quantified and combined in an overlapping pool design. Then, target genes are amplified from DNA pools and amplicons are combined to maximize throughput and increase likelihood of mutation detection during sequencing. Once sequence data is obtained, mutations are called using statistical approaches that weigh likelihood of rare mutations versus the probability of PCR and sequencing error.
You have full access to this open access chapter, Download chapter PDF
Similar content being viewed by others
Keywords
- Mutation discovery
- Reverse genetics
- TILLING (Targeting Induced Local Lesions in Genomes) by sequencing
1 Introduction
Targeting Induced Local Lesions in Genomes (TILLING) is a reverse genetics method that combines the generation of large mutant populations through traditional mutagenesis with the moderate- to high-throughput discovery of rare mutations. Originally described over a decade ago in Arabidopsis (McCallum et al. 2000) and Drosophila (Bentley et al. 2000), TILLING is a particularly attractive method for those working with species for which there are few, if any, genomics resources available (Comai and Henikoff 2006). The relative ease, low cost, and broad applicability of chemical mutagenesis enable rapid development of the requisite mutant populations in most species, in contrast to the generation of insertional mutants.
Traditional TILLING involves mutation detection based on enzymatic cleavage of mismatched sites in heteroduplexes formed after the annealing of PCR amplicons derived from genes of interest. This approach has been adapted for use by laboratories with limited resources (Jankowicz-Cieslak et al. 2012). However, discovery of mutations in pools of more than eight individuals is challenging, and once a putative mutation has been detected, Sanger sequencing of all individuals in the pool is needed to verify the mutation (McCallum et al. 2000; Bentley et al. 2000; Comai and Henikoff 2006; Jankowicz-Cieslak et al. 2012; Tsai et al. 2011). Because Sanger sequencing requires a large amount of template and is prone to sequencing errors, traditional TILLING can be slow and cost prohibitive. The advent of next-generation DNA sequencing (NGS), which is cheaper and faster than the classic Sanger method, provides new opportunities for mutant identification in TILLING populations (Tsai et al. 2011).
While traditional TILLING has been employed to great effect, the most direct approach for identifying mutations is to sequence putative mutant alleles and compare them to a reference sequence. Recent advances in ultrahigh-throughput sequencing and bioinformatics analyses have enabled the discovery of rare mutations based on massive parallel sequencing. Such an approach requires sufficient sequencing coverage to allow the discrimination of real mutations from changes introduced through sequencing errors. While the technology exists to detect single-nucleotide polymorphisms by whole-genome resequencing (Ossowski et al. 2010), the application of this approach to large mutant populations is still too costly for most species (Tsai et al. 2011). Instead, mutation discovery using massive parallel sequencing approaches have been focused on the gene space, where mutations are most likely to affect function. This can be accomplished by 3D DNA multiplexing followed by pooling of amplicons derived from genes of interest (Tsai et al. 2011, 2013; Rigola et al. 2009) or via the use of sequence capture methods (Gnirke et al. 2009; Fisher et al. 2011) to select for gene coding sequences (Porreca et al. 2007; Ng et al. 2009; Nijman et al. 2010). The latter of the two strategies represents a global or genome-wide approach to mutation discovery by focusing on all or most of the gene coding regions within a genome of interest (i.e., the exome). Indeed, exome sequencing has been successfully employed in animals (Ng et al. 2010; Ramos et al. 2012; Sun et al. 2012) and plants (Bolon et al. 2011; Henry et al. 2014) to discover rare mutations and could be extended to characterize large mutant populations such as those currently employed for TILLING, although the investment required likely limits this approach in plants to model and major economic species.
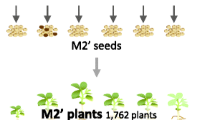
Sequencing of PCR amplicons represents a natural extension of the traditional TILLING method (Tsai et al. 2011; Rigola et al. 2009). An advantage of mutation detection using next-generation sequencing platforms is increased sensitivity, which enables discovery in more complex DNA pools (64× deep) than traditional TILLING (8× deep). This translates to more flexibility with regard to the pooling strategy, which in turn facilitates concurrent identification of mutations and the corresponding individuals. This approach increases the throughput of mutation discovery and speeds the identification and prioritization of putative mutants for in-depth functional studies. Here we describe a protocol for the TILLING by sequencing (TBS) of diploid individuals, which has been successfully employed in rice (Tsai et al. 2011) and tested in Arabidopsis, tomato, and Camelina (Tsai et al. 2011; unpublished data). In this version of TBS, populations of 512 M2 individuals are tridimensionally pooled and candidate genes are amplified and pooled, indexed and pooled, and sequenced (Fig. 20.1). Resulting sequencing reads are then processed and aligned to a reference genome for mutation discovery. By taking advantage of the high yield of next-generation sequencing platforms, it is possible to rapidly screen individuals with a higher degree of sensitivity. In addition, the availability of an array of sequencing platforms and bar coding allows users to employ multiple pooling schemes and flexibility with regard to the number of candidate genes that are screened. TBS can be easily adapted for mutation detection in other plant and animal species at a relatively low cost.
Targeted discovery of rare mutations in a rice mutant population via TILLING by sequencing. Diagram depicting TILLING by sequencing workflow starting with a mutant population and ending with mutant identification and confirmation. DNA is isolated, carefully quantified, and collapsed into pools, which are then amplified using PCR. Illumina libraries are prepared from fragmented PCR products and then sequenced. Putative mutations are confirmed through resequencing of deconvoluted pools
2 Materials
2.1 Genomic DNA Quantification
-
1.
Genomic DNA (see Note 1).
-
2.
TE buffer (10 mM Tris, 1 mM EDTA; pH 7.4).
-
3.
λ DNA (100 ng/μl).
-
4.
NanoDrop (Thermo Fisher Scientific) or similar instrument.
-
5.
SYBR Green I Nucleic Acid Stain (Lonza Cat. No. 50512). Store at −20 °C (see Note 2).
-
6.
PCR plates (standard 96-well).
-
7.
Adhesive plate seals for PCR (Applied Biosystems Cat. No. 4306311).
-
8.
Micropipettes (single and multichannel) and tips.
-
9.
1.7 ml microcentrifuge tubes.
-
10.
Costar® 96-well plates, flat bottom, black (Corning Cat. No. 3915).
-
11.
0.2 ml 8-strip PCR tubes with caps.
-
12.
Adhesive plate seals for storing samples in 96-well plates (Thermo Fisher Scientific Cat. No. AB-0580).
-
13.
Aluminum foil.
-
14.
Microplate reader (fluorescence).
2.2 DNA Pooling for PCR Preparation
-
1.
Diluted and normalized genomic DNA.
-
2.
PCR plate (standard 96-well).
-
3.
Multichannel micropipette and tips.
2.3 PCR, Quantification, and Cleanup of Target Amplicons
-
1.
Ex Taq DNA polymerase, Hot Start Version (Takara). Store at −20 °C (see Note 3).
-
2.
10× Ex Taq PCR buffer (supplied with Ex Taq). Store at −20 °C.
-
3.
Premixed 2.5 mM dNTPs (supplied with Ex Taq). Store at −20 °C.
-
4.
TE: 10 mM Tris–HCl 1 mM ethylenediaminetetraacetic acid (EDTA), pH 7.4.
-
5.
Left primer, Tm 67–73 °C, 100 μM in TE (see Note 4). Store at −80 °C.
-
6.
Right primer, Tm 67–73 °C, 100 μM in TE (see Note 4). Store at −80 °C.
-
7.
PCR plates (standard 96-well).
-
8.
Adhesive plate seals (Applied Biosystems Cat. No. 4306311).
-
9.
Thermocycler.
-
10.
1 % agarose/1× TAE gel and 1× TAE electrophoresis buffer.
-
11.
Standard agarose gel electrophoresis equipment.
-
12.
Agencourt AMPure XP beads (Beckman Coulter Scientific). Store at 4 °C.
-
13.
80 % ethanol (made fresh from 95 % stock solution).
-
14.
Elution buffer (10 mM Tris–HCl, pH 8.0).
-
15.
Magnetic separation device.
-
16.
SYBR Green I Nucleic Acid Stain (see Note 2).
-
17.
Costar® 96-well plates, flat bottom, black (Corning Cat. No. 3915).
-
18.
λ DNA (100 ng/μl).
-
19.
TE buffer (10 mM Tris, 1 mM EDTA; pH 7.4).
-
20.
0.2 ml 8-strip PCR tubes with caps.
2.4 Fragmentation of PCR Products
-
1.
NEBNext® dsDNA Fragmentase (New England Biolabs). Store at −20 °C.
-
2.
10× Fragmentase Buffer (New England Biolabs) supplied with NEBNext® dsDNA Fragmentase. Store at −20 °C.
-
3.
100× BSA (New England Biolabs) supplied with NEBNext® dsDNA Fragmentase. Store at −20 °C.
-
4.
0.5 M EDTA (pH 8.0).
-
5.
Agencourt AMPure XP beads.
-
6.
80 % ethanol (prepared fresh from 95 % stock solution).
-
7.
Elution buffer (EB; 10 mM Tris–HCl, pH 8.0).
2.5 NGS Library Preparation and Pooling
-
1.
NGS Library Preparation Kit (KAPA Biosystems). Store at −20 °C (see Note 5).
-
2.
NanoDrop (Thermo Fisher Scientific) or similar instrument.
-
3.
Agencourt AMPure XP beads.
-
4.
80 % ethanol (prepared fresh).
-
5.
Elution buffer (10 mM Tris–HCl, pH 8.0).
-
6.
Indexed adapters (100 μM in TE). Stored at −20 °C (see Note 6).
-
7.
Adapter mixture: 5 μM premixed adapters (universal adapter + indexed adapter), diluted from 100 μM stocks with distilled deionized water. Stored at 4 °C (short term) or −20 °C (long term).
-
8.
Left primer for adapter amplification, 100 μM in TE. 5′-AATGATACGGCGACCACCGAGATCTACAC-3′. Stored at −20 °C.
-
9.
Right primer for adapter amplification, 100 μM in TE. 5′-CAAGCAGAAGACGGCATACGAGAT-3′. Stored at −20 °C.
-
10.
Library enrichment primer mixture: 5 μM left primer + right primer (diluted from 100 μM stocks with distilled deionized water).
2.6 Library Sequencing
-
1.
Pooled, enriched libraries.
-
2.
Qubit fluorometer (Thermo Fisher Scientific).
-
3.
Qubit dsDNA High Sensitivity Assay Kit (Thermo Fisher Scientific).
-
4.
Clear fluorometry tubes for DNA quantification.
-
5.
Bioanalyzer (Agilent) for library size estimates.
2.7 Bioinformatics Analysis
-
1.
1 TB hard drive for compressed raw read storage.
-
2.
2 TB storage space for read processing and data analysis.
-
3.
Minimal memory required = ~8 Gb.
2.8 Mutation Confirmation
-
1.
Plant growth supplies.
-
2.
Genomic DNA from mutant plants.
3 Methods
3.1 Genomic DNA Quantification
DNA quantification is perhaps the most important part of TILLING method. If the input the DNA is variable, the ability to detect mutations will be diminished. Fluorescence-based methods for DNA quantification are recommended because they scale well to hundreds of individuals, but alternate methods are available (see Note 7).
-
1.
Prepare λ DNA standard by diluting 100 ng stock with TE in serial dilution.
-
(a)
Vortex the 100 ng/μl λ DNA and spin down. Then, identify the concentration by recording the average of three NanoDrop (see Note 8) readings.
Concentration (ng/μl) = _______________
If stock is not 100 ng, adjust standard concentrations accordingly.
-
(b)
Prepare the λ DNA standard in 0.2 ml PCR tubes (8 or 12-tube strips may be used for convenience).
-
(c)
Pipette Std.1 into first tube.
-
(d)
Transfer 30 μl to the next well and add 20 μl 1XTE and mix well. This is the first dilution step.
-
(e)
Prior to starting subsequent dilution, vortex and spin down.
-
(a)
Std. | Conc. | Dilution |
|---|---|---|
1 | 100 | 50 μl starting concentration |
2 | 60 | 30 μl Std.1 + 20 μl 1XTE |
3 | 36 | 30 μl Std.2 + 20 μl 1XTE |
4 | 21.6 | 30 μl Std.3 + 20 μl 1XTE |
5 | 12.96 | 30 μl Std.4 + 20 μl 1XTE |
6 | 7.78 | 30 μl Std.5 + 20 μl 1XTE |
7 | 4.67 | 30 μl Std.6 + 20 μl 1XTE |
8 | 2.8 | 30 μl Std.7 + 20 μl 1XTE |
-
2.
In a light-protected tube or reagent reservoir, prepare SYBR Green I master mix by combining 200 μl TE with 0.08 μl SYBR Green I for each reaction, and mix well (see Note 9). Protect mixture from light to prevent loss of fluorescence signal.
-
3.
Pipette 200 μl 1× SYBR Green I master mix into each well of a black Costar plate (see Note 10).
-
4.
Transfer 2 μl λ DNA standard or 2 μl unknown DNA (see Note 11) to wells containing 1× SYBR Green I master mix. Mix well by pipetting up and down.
-
5.
Cover plates with sealing film and aluminum foil.
-
6.
Shake for 2.5 min at 55 rpm on a plate shaker (or vortex with platform attachment) and then spin down. Store the plates in the dark at 4 °C prior to quantification.
-
7.
Measure fluorescence with a microplate fluorescence reader according to manufacturer’s protocol to determine DNA concentration.
-
8.
Once DNA is quantified with a plate reader, calculate DNA concentration and appropriate normalization. The final concentration should be equal to the most dilute DNA sample and consistent across plates (if at all possible). Final concentration should not be <2.0 ng/μl (see Note 12).
3.2 Three-Dimensional Pooling of Genomic DNAs
-
1.
Layout eight “pre-pool” 96-well PCR plates, which will contain diluted DNA (see Sect. 20.3.1, Step 8) to be used for constructing an 8 × 8 pooled array.
-
2.
Transfer at least 35 μl of diluted DNA per sample to wells in columns 1–8 and rows A–H of the pre-pool plates (see Note 13). Label two sets of 24 microfuge tubes (1.7 ml), which will contain the master pools. Master pools are labeled by row (R1–R8), column (C1–C8), and plate (P1–P8). Collapse pre-pool plates into master pools (see Fig. 20.2 and Note 14). Using multichannel pipette is probably the most efficient way to collapse pre-pool plates. It also reduces probability of sample swaps. To do this, collapse pre-pool plates in another PCR plate and then transfer to a microfuge tube. All pooling should be done in duplicate. Master pools can be kept at 4 °C for short-term and −20 °C/−80 °C for long-term storage.). For example:
-
(a)
R3 master pool (horizontal pool):
-
Combine 5 μl DNA from row 3 (wells C1–C8) from pre-pool plates #1–8. This will yield a pool of 64 individuals.
-
-
(b)
C1 master pool (vertical pool):
-
Combine 5 μl DNA from column 1 (wells A1–H1) from pre-pool plates #1–8. This will yield a pool of 64 individuals.
-
-
(c)
P4 master pool (plate pool):
-
Combine 5 μl DNA from all individuals on pre-pool plate #4 (row wells A–H and column wells 1–8). This will yield a pool of 64 individuals.
-
-
(a)
-
3.
Quantify master pools with a NanoDrop or with a similar method and proceed to PCR amplification.
Three-dimensional pooling facilitates simultaneous mutation detection and identification of individual mutants harboring the mutations. Mutant individuals are arrayed on eight pre-pool plates with one individual in each well. Plates are then pooled in three dimensions. Each individual is present in three pools (a row pool, R; a column pool, C; and a plate pool, P) and each pool contains 64 individuals
3.3 PCR and Pooling of TILLING Targets
-
1.
Program a thermocycler with the following amplification profile (see Note 15):
-
95 °C for 2 min (initial denaturation)
-
Eight cycles of: | 73 °C for 30 s. |
(Increments of −1 °C/cycle, ramp to 72 °C at 0.5 °C/s) | |
72° for 1 min | |
25 cycles of: | 94 °C for 20 s |
65 °C for 30 s (ramp to 72 °C at 0.5 °C/s). | |
72 °C for 1 min |
-
2.
Set up the following PCR reaction on ice (see Note 16):
DNA (1.5 ng) and water | 22.65 μl |
10× Ex Taq buffer | 3.00 μl |
2.5 mM dNTPs | 2.40 μl |
5 μM premixed primers | 1.80 μl |
Ex Taq HS enzyme | 0.15 μl |
Final volume | 30.0 μl |
-
3.
To check reaction efficiency, run 1 μl of the 30 μl PCR on 1 % agarose/1× TAE gel (see Note 17).
-
4.
Follow DNA quantification protocol described above to quantify PCR products (see subheading 20.3.1, and Note 18), and calculate PCR sample normalization (see Note 19).
-
5.
Normalize the PCR product.
-
6.
Calculate the average amount (in ng) of each of the PCR products needed to obtain 500 ng of pooled PCR DNA for sequencing library preparation by dividing 500 ng by the number of PCR products (see Note 20).
-
7.
For each target, calculate the size factor by dividing the PCR fragment length by the mean fragment length (see Note 21).
-
8.
For each target, calculate the input amount (in ng) needed by multiplying the average amount (from step 6) by the size factor (from step 7). This is the amount you will add as input DNA.
-
9.
For each target, calculate input volume by dividing the final volume (from step 8) by the normalized concentration.
-
10.
Pool PCR products according to input volume. Clean PCR pool with an equal volume of AMPure XP beads (1:1) and elute in 52 μl water or elution buffer (see Note 22).
3.4 Fragmentation of PCR Products
-
1.
Set up fragmentation reactions on ice (see Note 23).
DNA and water | 51.4 μl |
10× Fragmentase buffer | 6.0 μl |
100× BSA | 0.6 μl |
dsDNA fragmentase | 2.0 μl |
Final volume | 60.0 μl |
-
2.
Incubate at 37 °C for ~20 min (see Note 24).
-
3.
Remove 2 μl of the 60 μl reaction and store the remainder at −20 °C. Check reaction efficiency by running the 2 μl sample on a 1 % agarose/1× TAE gel with a 1 kb DNA ladder.
-
4.
If most DNA fragments are between 300 and 600 bp, then proceed to step 5. If there are many large fragments (>800 bp), place the frozen reaction back at 37 °C for another 5–10 min and check again on gel.
-
5.
Add 5 μl 0.5 M EDTA and mix well to stop the reaction.
-
6.
Clean DNA by mixing 60 μl AMPure XP beads (1:1 v/v) with the fragmentation reaction and follow the manufacturer’s protocol for AMPure cleanup of PCR reactions (see Note 22).
-
7.
Elute DNA with 50 μl elution buffer.
3.5 NGS Library Preparation and Pooling
-
1.
Follow protocol outlined in the KAPA Technical Data Sheet (see Note 25). All reactions are performed according to the manufacturer’s specifications except 1 μl premixed 5 μM adapters, and 4 μl of water is added to ligation reaction (see Note 26).
-
2.
Determine concentration of each of the enriched libraries by analyzing 1 μl using a NanoDrop or Qubit. If some libraries are a lot more concentrated than the mean across all other samples, dilute accordingly, and then re-quantify the dilution.
-
3.
Assuming relatively similar fragment size distribution, pool samples 1:1 based on the DNA amount which is derived from the sample concentration obtained from SYBR Green 1 quantification (see Note 27).
3.6 Sequencing of NGS Libraries
The NGS libraries constructed from the PCR pools in this protocol are for Illumina NGS platforms (e.g., HiSeq 3000 or 2500) and are typically submitted to sequencing core facilities at universities or research institutions or to commercial sequencing labs. The protocols for submission, handling, and analysis of samples are dependent on the facility. A general outline for sample submission can be found below.
-
1.
Determine purity of pooled libraries through fluorometry, for example, Qubit quantification (see Note 28).
-
2.
Assess quality of libraries through qPCR, such as the KAPA Library Quantification Kit (see Note 29). This will yield an estimate of cluster forming capacity and thus predicted read output from Illumina sequencing.
-
3.
Confirm library size with Bioanalyzer (Agilent), which can quantify the size of small samples of DNA (see Note 30).
-
4.
Select desired number of reads/sequencing lanes. This is dependent on (1) the number of TILLING targets, (2) size of TILLING targets (before library preparation), (3) read length, and (4) predicted number of sequencing reads (see Note 31).
3.7 Computational Analysis of Sequence Data
A bioinformatics pipeline is required to process the large volume of sequence data that is generated and to effectively discriminate between real mutations and background “noise.” This is complicated by the fact that the intensity of the mutation “signal” and the background noise may vary greatly as a result of the sequence coverage and the location of the mutation (see Note 32). The specifics of the bioinformatics analysis are beyond the scope of this chapter and have been described in detail elsewhere (see Note 32). The general steps for mutation discovery are as follows:
-
1.
Obtain Illumina raw sequence reads (see Note 33).
-
2.
Quality filter the sequence reads and trim to remove adapter sequences.
-
3.
Identify samples according to index and de-multiplex (see Note 34).
-
4.
Align sequences to reference using Burrows-Wheeler Aligner (BWA) software (Li and Durbin 2009).
-
5.
Identify and parse sequence changes from BWA output.
-
6.
Compare frequency of the sequence changes across libraries.
-
7.
Derive a probability score for each sequence change based on quality scores and other parameters (see Note 35).
3.8 Confirmation of Mutations and Initial Characterization of Mutants
Putative mutations identified through the bioinformatics pipeline are prioritized for further analysis based on their nature (e.g., nonsense, missense) and location (e.g., exon, intron, splice site, untranslated regions).
-
1.
Obtain the M3 seeds corresponding to the M2 individual identified as harboring a mutation of interest. Evaluate seed and plant phenotypes as warranted.
-
2.
Harvest tissue and prepare DNA from M3 plants.
-
3.
Identify M3 individuals harboring the mutation of interest by Sanger sequencing or, if possible, restriction digestion (see Note 36).
-
4.
Cross M3 individuals with wild-type and other mutants to produce materials for further analysis (see Note 37).
4 Notes
-
1.
Any extraction protocol that yields DNA of high molecular weight (≥15 kb) and good purity (A260/A280 ratio of ≥1.8). We have used both homemade reagents (e.g., potassium acetate–sodium dodecyl sulfate method) and commercial kits (e.g., Qiagen DNeasy). Total genomic DNA (including chloroplast and mitochondrial genomes) is routinely used.
-
2.
SYBR Green I Stain can be obtained from many suppliers. Aliquots should be made to avoid freeze thawing. Because of light-sensitive and fluorescence decay, the stock solution as well as any working stocks should be kept covered with foil at all times. It should be thawed to room temperate for 30 min before use. As with any reagent that binds to nucleic acids, it should be handled with care and disposed of according to your institution’s hazardous waste policy. Additional safety information can be obtained from the Material Safety Data Sheet.
-
3.
Takara Ex Taq HS DNA Polymerase works well for most TILLING applications. However, for difficult templates, alternate polymerases may be considered. To reduce false positives due to polymerase error, an enzyme with a low error rate and proofreading ability is advisable. Phusion® High-Fidelity DNA Polymerase (New England Biolabs) is a likely candidate because of its robust performance. When considering alternatives to Ex Taq, additional time may be needed for protocol optimization.
-
4.
Primers should be designed with Primer3 or similar software and ideally sit in intergenic regions flanking exons. Optimal primer length is 27 bp (range of 20–30 bp) and primer temperature of 67 °C–73 °C (Till et al. 2007). Product size should be between 1000 bp and 2000 bp, with an optimal size of 1500 bp. Larger amplicons are possible, but it will be necessary to perform additional fragmentation trials.
-
5.
Next-generation sequencing (NGS) library kits are available from many commercial sources. Homemade kits consisting of components obtained from various sources may also be used. We have found the convenience and reliability of commercial kits to outweigh the possible cost savings of assembling individual reagents. While it is possible to prepare PCR fragments for sequencing with a homemade DNA-seq kit, it is more efficient with a kit. There are many library preparation kits available on the market which contain all enzymes required for library construction, and reactions are performed in the presence of magnetic beads, thus cutting down on the cost of multiple AMPure reactions.
-
6.
These adapters serve multiple functions. They are used for amplification of the libraries, they included specific barcodes (usually 6 or 8 bp long) that identified the samples, and they bind to the sequencing flow cell. Premade indexed adapters can be ordered from many companies including Illumina (San Diego, CA, USA), Bioo Scientific (Austin, TX, USA), and New England Biolabs (Ipswich, MA, USA), or homemade adapters can be designed. The “TruSeq” Illumina single index adapter system has a universal adapter and a 6- or 8-bp indexed adapter. The adaptors are complementary in a 12-base region; binding in this region creates the “Y-shaped” adaptor—a short region of complementarity with long mismatched arms. Barcodes should be unique to each pool and differ at a minimum of 2 bps (for 6-bp barcodes) or 3 bps (for 8-bp barcodes), to easily distinguish between pools at the bioinformatic analysis stage.
-
7.
If a microplate reader capable of fluorescence detection is not available, gel electrophoresis with λ DNA standard has been effectively used to quantify DNA for TILLING (Till et al. 2003; Cooper et al. 2008).
-
8.
Any method for quantifying the λ DNA can be used here.
-
9.
SYBR Green I dye master mix is prepared fresh prior to quantification. Work quickly and keep all reagents in the dark! Make 1× SYBR Green I dye master mix by combining the following:
TE (ml) = 0.2× (# reactions) = ___________
SYBR Green I (μl) = 0.08× (# reactions) = ___________
Make sure to include at least 24 standard reactions, eight blank reactions, extra for pipetting error, etc.
-
10.
Corning Costar® black plates are used for fluorescence-based assays as the absorbed light, thus reducing background and cross talk. Pipette 200 μl 1× SYBR Green I dye mixture into each well of labeled quantification plate using a P200 or P1000 multichannel pipette. Cover with aluminum foil until ready for use.
-
11.
Add 2 μl of DNA standard to SYBR Green I mixture. Pipette up and down a few times to make sure no liquid is retained in tip. Standards should be done in triplicates and are usually place in A1–H1, A2–H2, and A3–H3. Leave wells A4–H4 blank. Add 2 μl unknown DNA to SYBR Green I mixture. Unknowns can be should be aliquoted as singlets. Pipette up and down a few times to make sure no liquid is retained in tip. If quantifying a large number of samples, plates should be covered with foil to protect from light and placed at 4 °C while the remaining 1× SYBR Green I mix is distributed.
-
12.
DNA samples which have an initial concentration below 2 ng/μl should be excluded, or more DNA should be extracted. Very concentrated templates are also not ideal. Calculating appropriate initial concentration is crucial to the success of this experiment. Take extra care when pipetting and consider repeating quantification if any of the following occur: (1) Standard curve has low R-value (less than 0.95). If this is the case, discard experiment. (2) DNA concentration is not within range of curve. If initial concentration is >100 ng, dilute DNA and re-quantify. If initial concentration is <2 ng, either (a) DNA was not added or (b) DNA extraction needs to be redone. (3) Initial concentration calculated for standards is far off from the expected values. If this is observed, quantify (e.g., NanoDrop) all standards and run on gel (λ DNA could be degraded).
-
13.
This amount depends on starting concentration of dilutions. DNA from all samples must be the same concentration. If it is not standardized, dilute accordingly. A sufficient number of input DNA molecules are needed for accurate mutation calling. Studies in diploid rice estimate that 24 copies of DNA per allele ensure coverage of the mutant allele and reduce copy number variants (Tsai et al. 2011). Having fewer copies of DNA in pools increases the probability of having no coverage of an individual allele and increases likelihood of false mutation calls due to polymerase-based error (Tsai et al. 2011; Akbari et al. 2005). For a 64-individual pool, approximately 1.5 ng template is needed for PCR. Diploid rice has 2C value of approximately 1.0 pg (Johnston et al. 1999), therefore 0.5 ng total for each dimensional pool (Tsai et al. 2011). After preparing master pools, pre-pool plates can be heat sealed and stored at −20 °C.
-
14.
It is good to include control(s) to ensure pooling has been done correctly. Include at least one DNA sample with a known mutation in a gene that you are amplifying on a pre-pool plate. Pooling is perhaps the most difficult/error-prone step of TBS pipeline. Use caution when pipetting (i.e., make sure you are always transferring the same amount of DNA).
-
15.
Because TILLING primers are likely to have high annealing temperatures (due to length and GC content), the use of a touchdown thermocycling program is employed to prevent non-specific annealing.
-
16.
It is very important to have specific amplification for mutation detection. The use of ice and a hot start taq polymerase significantly reduces non-specific priming. To increase efficiency, PCR reactions are performed in a 96-well assay. PCR master mix for a 96-well plate (including 5 % extra of each reagent and not including template DNA): 303 μl 10× Ex Taq buffer, 242.4 μl of 2.5 mM dNTP mixture, 181.8 μl of 10 μM primer mix, and 15.2 μl of Takara Ex Taq HS enzyme (0.75 unit/reaction).
-
17.
Failed reactions should be set up again in duplicate or triplicate.
-
18.
Do not use a NanoDrop to determine sample concentration of PCR reactions. The presence of dNTPs, polymerase, etc., will skew results.
-
19.
While it is necessary for all PCR from a particular primer pair to be of the same concentration, PCR products from different genes do not need to be standardized. As with DNA normalization, the final dilution will most likely be determined by the concentration of the most dilute sample on the plate. Dilute PCR products by adding appropriate volume of TE.
-
20.
Larger fragments need more input DNA because mutation detection requires an even representation of each base in the sequencing space. For rice TILLING, 500 ng–1 μg of pooled PCR product is sufficient for library preparation and mutation discovery. The current protocol is optimized for 500 ng of pooled PCR product with an average size of ~1.5 kb.
-
21.
For example, to sequence five TILLING fragments of the same size, you would need 100 ng DNA for each fragment. This amount should be adjusted according to fragment size, i.e., larger fragments require a proportionally greater amount of input DNA.
-
22.
Agencourt AMPure XP beads can be stored at 4 °C for approximately 1 year. Beads settle over time, so it is necessary to fully resuspend beads prior to use. Observe all of the manufacturer’s guidelines for handling beads.
-
23.
NEBNext® Fragmentase enzyme will digest up to 3 μg DNA. However, enzyme amount and incubation time will need to be adjusted. Take special care when working with Fragmentase. The enzyme needs to be vortexed prior to reaction setup. It is important to keep all samples on ice (or at −20 °C for longer periods), thus reducing enzymatic activity and avoiding undesirable digestion. In addition, it appears that digestion can be highly variable depending on template or Fragmentase batch. Optimize enzyme amount (try between 1 and 3 μl) and digestion time (20–45 min) on a few samples prior to scaling to all pools. DNA can also be mechanically fragmented by sonication, for example, with a Covaris Focused-ultrasonicator. This yields consistent shearing with reduced template bias and can be preferable to chemical fragmentation because it does not depend on template concentration and yields a more uniform fragment distribution.
-
24.
This time may be shorter or longer and is determined by visualizing fragment size on a gel.
-
25.
The KAPA data sheet has a lot of good information regarding handling of reagents. For example, AMPure beads and PEG/SPRI solution must be equilibrated to room temperature prior to use. It is strongly suggested that it is read in entirety.
-
26.
Approximately eight cycles should be sufficient for enrichment although new kits may need only 2-4 cycles or can be used PCR-free. Run 5 μl of 50 μl of enrichment PCR on an agarose gel to confirm (while sample is stored at 4 °C). Reaction can be returned to thermocycler for another 2–4 cycles if needed. Do not exceed 12 cycles of enrichment. After final PCR cleanup, run 2 μl of 20 μl on 2 % gel to make sure that library is primer and adapter-free. This can also be confirmed with a Bioanalyzer trace.
-
27.
DNA concentration can be obtained through SYBR Green 1 (see Methods Sect. 20.3.1). NanoDrop may also be used but does not yield as reliable results. Most sequencing facilities will not need very much input DNA (e.g., 10 μl of 10 nM). Therefore do not pool all of your samples. Start with 1–2 μl of most concentrated sample and adjust amount of others accordingly.
-
28.
Input DNA purity should have an approximate 260/230 >2.0 and 260/230 between 1.8 and 2.0.
-
29.
Illumina sequencing facilities often offer library quantification services as part of sequencing. Therefore, it is usually not necessary to perform library quantification prior to library submission.
-
30.
If 150 bp paired-end reads are obtained, the average insert size for sequencing should be approximately 350 bp (not including adaptors).
-
31.
On the HiSeq 2500, assuming 150PE reads, approximately 1024 individuals, with 20 (1500 bp) targets can be screened in a single HiSeq lane.
-
32.
Details of the bioinformatics pipeline can be found in Tsai et al. (2011, 2013) and Missirian et al. (2011). Like sequencing, most labs may not have the necessary personnel and resources to conduct the bioinformatics analyses completely in-house. Sequencing core facilities are typically associated with bioinformatics analysis core groups that can provide support and some infrastructure to facilitate more efficient processing and analysis of sequence data.
-
33.
Sequence reads are provided as compressed FASTQ format files that contain both the reads that passed quality filters and those that did not. It is recommended that alignment be performed on a computer cluster as memory requirements for alignment and mutation identification exceed that which can be handled on a personal computer.
-
34.
Each quality trimmed read file (sequenced libraries from row, column, and plate pools) is placed in its own directory. For example, reads from first-row pool would be placed in directory labeled “T1R1.”
-
35.
Probability scores are assigned using Coverage Aware Mutation Calling Using Bayesian Analysis (CAMBa; available at http://web.cs.ucdavis.edu/~filkov/CAMBa/), a software program that estimates the probability that a mutation is falsely identified with a Ft score, the mean-centered log of the posterior probability of mutation (Tsai et al. 2011; Missirian et al. 2011). CAMBa uses the overlaps resulting from the tridimensional library pooling strategy to identify the mutations and the corresponding mutant individuals simultaneously.
-
36.
Use original TILLING primers or newly designed primers closer to the location of the mutation to amplify and sequence to confirm mutation and identify individuals that are heterozygous and homozygous at the mutation. If the mutation is predicted to create or remove a restriction enzyme recognition site, it may be more efficient and cost effective to perform restriction digest of PCR products.
-
37.
Generally backcrossing of mutants to the wild-type cultivar from which the mutant population was derived is performed to eliminate mutations that may confound characterization. It may be necessary to backcross two or more times. It may also be possible and useful to evaluate the M3 plants directly depending on the nature of the mutant phenotype. It is recommended that approximately 20–25 M3 plants should be screened for stable mutation recovery.
References
Akbari M, Hansen MD, Halgunset J, Skorpen F, Krokan HE (2005) Low copy number DNA template can render polymerase chain reaction error prone in a sequence-dependent manner. J Mol Diagn 7:36–39
Bentley A, MacLennan B, Calvo J, Dearolf CR (2000) Targeted recovery of mutations in Drosophila. Genetics 156:1169–1173
Bolon YT, Haun WJ, Xu WW, Grant D, Stacey MG, Nelson RT, Gerhardt DJ, Jeddeloh JA, Stacey G, Muehlbauer GJ, Orf JH, Naeve SL, Stupar RM, Vance CP (2011) Phenotypic and genomic analyses of a fast neutron mutant population resource in soybean. Plant Physiol 156:240–253
Comai L, Henikoff S (2006) TILLING: practical single-nucleotide mutation discovery. Plant J 45:684–694
Cooper JL, Greene EA, Till BJ, Codomo CA, Wakimoto BT, Henikoff S (2008) Retention of induced mutations in a Drosophila reverse-genetic resource. Genetics 180:661–667
Fisher S, Barry A, Abreu J, Minie B, Nolan J, Delorey TM, Young G, Fennell TJ, Allen A, Ambrogio L, Berlin AM, Blumenstiel B, Cibulskis K, Friedrich D, Johnson R, Juhn F, Reilly B, Shammas R, Stalker J, Sykes SM, Thompson J, Walsh J, Zimmer A, Zwirko Z, Gabriel S, Nicol R, Nusbaum C (2011) A scalable, fully automated process for construction of sequence-ready human exome targeted capture libraries. Genome Biol 12:R1
Gnirke A, Melnikov A, Maguire J, Rogov P, LeProust EM, Brockman W, Fennell T, Giannoukos G, Fisher S, Russ C, Gabriel S, Jaffe DB, Lander ES, Nusbaum C (2009) Solution hybrid selection with ultra-long oligonucleotides for massively parallel targeted sequencing. Nat Biotechnol 27:182–189
Henry IM, Nagalakshmi U, Lieberman MC, Ngo KJ, Krasileva KV, Vasquez-Gross H, Akhunova A, Akhunov E, Dubcovsky J, Tai TH, Comai L (2014) Efficient genome-wide detection and cataloging of EMS-induced mutations using exome capture and next-generation sequencing. Plant Cell 26:1382–1397
Jankowicz-Cieslak J, Huynh OA, Dussoruth B, Saraye B, Till BJ (2012) Low cost mutation discovery methods suitable for developing countries. Sci MED 3:245–249
Johnston JS, Bennett MD, Rayburn AL, Galbraith DW, Price HJ (1999) Reference standards for determination of DNA content of plant nuclei. Am J Bot 86:609–613
Li H, Durbin R (2009) Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics 25:1754–1760
McCallum CM, Comai L, Greene EA, Henikoff S (2000) Targeted screening for induced mutations. Nat Biotechnol 18:455–457
Missirian V, Comai L, Filkov V (2011) Statistical mutation calling from sequenced overlapping DNA pools in TILLING experiments. BMC Bioinf 12:287
Ng SB, Turner EH, Robertson PD, Flygare SD, Bigham AW, Lee C, Shaffer T, Wong M, Bhattacharjee A, Eichler EE, Bamshad M, Nickerson DA, Shendure J (2009) Targeted capture and massively parallel sequencing of 12 human exomes. Nature 461:272–276
Ng SB, Buckingham KJ, Lee C, Bigham AW, Tabor HK, Dent KM, Huff CD, Shannon PT, Jabs EW, Nickerson DA, Shendure J, Bamshad MJ (2010) Exome sequencing identifies the cause of a mendelian disorder. Nat Genet 42:30–35
Nijman IJ, Mokry M, van Boxtel R, Toonen P, de Bruijn E, Cuppen E (2010) Mutation discovery by targeted genomic enrichment of multiplexed barcoded samples. Nat Methods 7:913–915
Ossowski S, Schneeberger K, Lucas-Lledó JI, Warthmann N, Clark RM, Shaw RG, Weigel D, Lynch M (2010) The rate and molecular spectrum of spontaneous mutations in Arabidopsis thaliana. Science 327:92–94
Porreca GJ, Zhang K, Li JB, Xie B, Austin D, Vassallo SL, LeProust EM, Peck BJ, Emig CJ, Dahl F, Gao Y, Church GM, Shendure J (2007) Multiplex amplification of large sets of human exons. Nat Methods 4:931–936
Ramos E, Levinson BT, Chasnoff S, Hughes A, Young AL, Thornton K, Li A, Vallania FL, Province M, Druley TE (2012) Population-based rare variant detection via pooled exome or custom hybridization capture with or without individual indexing. BMC Genomics 13:683
Rigola D, van Oeveren J, Janssen A, Bonné A, Schneiders H, van der Poel HJ, van Orsouw NJ, Hogers RC, de Both MT, van Eijk MJ (2009) High throughput detection of induced mutations and natural variation using KeyPoint technology. PLoS One 4:e4761
Sun M, Mondal K, Patel V, Horner VL, Long AB, Cutler DJ, Caspary T, Zwick ME (2012) Multiplex chromosomal exome sequencing accelerates identification of ENU-induced mutations in the mouse. G3 2:143–150
Till BJ, Reynolds SH, Greene EA, Codomo CA, Enns LC, Johnson JE, Burtner C, Odden AR, Young K, Taylor NE, Henikoff JG, Comai L, Henikoff S (2003) Large-scale discovery of induced point mutations with high-throughput TILLING. Genome Res 13:524–530
Till BJ, Cooper J, Tai TH, Colowit P, Greene EA, Henikoff S, Comai L (2007) Discovery of chemically induced mutations in rice by TILLING. BMC Plant Biol 7:19
Tsai H, Howell T, Nitcher R, Missirian V, Watson B, Ngo KJ, Lieberman M, Fass J, Uauy C, Tran RK, Khan AA, Filkov V, Tai TH, Dubcovsky J, Comai L (2011) Discovery of rare mutations in populations: TILLING by sequencing. Plant Physiol 156:1257–1268
Tsai H, Missirian V, Ngo KJ, Tran RK, Chan SR, Sundaresan V, Comai L (2013) Production of a high-efficiency TILLING population through polyploidization. Plant Physiol 161:1604–1614
Acknowledgments
The methods described here were developed with the support from the National Science Foundation Plant Genome Program (Plant Genome award no. DBI–0822383) to L.C. and the US Department of Agriculture, Agricultural Research Service CRIS Project 5306-21000-017/021-00D to T.H.T.
Author information
Authors and Affiliations
Corresponding author
Editor information
Editors and Affiliations
Rights and permissions
Open Access This chapter is distributed under the terms of the Creative Commons Attribution-Noncommercial 2.5 License (http://creativecommons.org/licenses/by-nc/2.5/) which permits any noncommercial use, distribution, and reproduction in any medium, provided the original author(s) and source are credited.
The images or other third party material in this chapter are included in the work’s Creative Commons license, unless indicated otherwise in the credit line; if such material is not included in the work’s Creative Commons license and the respective action is not permitted by statutory regulation, users will need to obtain permission from the license holder to duplicate, adapt or reproduce the material.
Copyright information
© 2017 International Atomic Energy Agency
About this chapter
Cite this chapter
Burkart-Waco, D., Tsai, H., Ngo, K., Henry, I.M., Comai, L., Tai, T.H. (2017). Next-Generation Sequencing for Targeted Discovery of Rare Mutations in Rice. In: Jankowicz-Cieslak, J., Tai, T., Kumlehn, J., Till, B. (eds) Biotechnologies for Plant Mutation Breeding. Springer, Cham. https://doi.org/10.1007/978-3-319-45021-6_20
Download citation
DOI: https://doi.org/10.1007/978-3-319-45021-6_20
Published:
Publisher Name: Springer, Cham
Print ISBN: 978-3-319-45019-3
Online ISBN: 978-3-319-45021-6
eBook Packages: Biomedical and Life SciencesBiomedical and Life Sciences (R0)