Abstract
MIMIC is a Medical Information Mart for Intensive Care and consists of several comprehensive data streams in the intensive care environment, in high levels of richness and detail, supporting complex signal processing and clinical querying that could permit early detection of complex problems, provide useful guidance on therapeutic interventions, and ultimately lead to improved patient outcomes.
You have full access to this open access chapter, Download chapter PDF
Similar content being viewed by others
Keywords
FormalPara Take Home Messages-
MIMIC is a Medical Information Mart for Intensive Care and consists of several comprehensive data streams in the intensive care environment, in high levels of richness and detail, supporting complex signal processing and clinical querying that could permit early detection of complex problems, provide useful guidance on therapeutic interventions, and ultimately lead to improved patient outcomes.
-
This complicated effort required a committed and coordinated collaboration across academic, industry, and clinical institutions to provide a radically open access data platform accessible by researchers around the world.
1 The Vision
Patients in hospital intensive care units (ICUs) are physiologically fragile and unstable, generally have life-threatening conditions, and require close monitoring and rapid therapeutic interventions. They are connected to an array of equipment and monitors, and are carefully attended by the clinical staff. Staggering amounts of data are collected daily on each patient in an ICU: multi-channel waveform data sampled hundreds of times each second, vital sign time series updated each second or minute, alarms and alerts, lab results, imaging results, records of medication and fluid administration, staff notes and more. In early 2000, our group at the Laboratory of Computational Physiology at MIT recognized that the richness and detail of the collected data opened the feasibility of creating a new generation of monitoring systems to track the physiologic state of the patient, employing the power of modern signal processing, pattern recognition, computational modeling, and knowledge-based clinical reasoning. In the long term, we hoped to design monitoring systems that not only synthesized and reported all relevant measurements to clinicians, but also formed pathophysiologic hypotheses that best explained the observed data. Such systems would permit early detection of complex problems, provide useful guidance on therapeutic interventions, and ultimately lead to improved patient outcomes.
It was also clear that although petabytes of data are captured daily during care delivery in the country’s ICUs, most of these data were not being used to generate evidence or to discover new knowledge. The challenge, therefore, was to employ existing technology to collect, archive and organize finely detailed ICU data, resulting in a research resource of enormous potential to create new clinical knowledge, new decision support tools, and new ICU technology. We proposed to develop and make public a “substantial and representative” database gathered from complex medical and surgical ICU patients.
2 Data Acquisition
In 2003, with colleagues from academia (Massachusetts Institute of Technology), industry (Philips Medical Systems), and clinical medicine (Beth Israel Deaconess Medical Center, BIDMC) we received NIH (National Institutes of Health) funding to launch the project “Integrating Signals, Models and Reasoning in Critical Care”, a major goal of which was to build a massive critical care research database. The study was approved by the Institutional Review Boards of BIDMC (Boston, MA) and MIT (Cambridge, MA). The requirement for individual patient consent was waived because the study would not impact clinical care and all protected health information was to be de-identified.
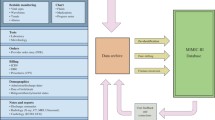
We set out to collect comprehensive clinical and physiologic data from all ICU patients admitted to the multiple adult medical and surgical ICUs of our hospital (BIDMC). Each patient record began at ICU admission and ended at final discharge from the hospital. The data acquisition process was continuous and invisible to staff. It did not impact the care of patients or methods of monitoring. Three categories of data were collected: clinical data, which were aggregated from ICU information systems and hospital archives; high-resolution physiological data (waveforms and time series of vital signs and alarms obtained from bedside monitors); and death data from Social Security Administration Death Master Files (See Fig. 5.1).
2.1 Clinical Data
Bedside clinical data were downloaded from archived data files of the CareVue Clinical Information System (Philips Healthcare, Andover, MA) used in the ICUs. Additional clinical data were obtained from the hospital’s extensive digital archives. The data classes included:
-
Patient demographics
-
Hospital administrative data: admission/discharge/death dates, room tracking, billing codes, etc.
-
Physiologic: hourly vital signs, clinical severity scores, ventilator settings, etc.
-
Medications: IV medications, physician orders
-
Lab tests: chemistry, hematology, ABGs, microbiology, etc.
-
Fluid balance data
-
Notes and reports: Discharge summaries; progress notes; ECG, imaging and echo reports.
2.2 Physiological Data
Physiological data were obtained with the technical assistance of the monitoring system vendor. Patient monitors were located at every ICU patient bed. Each monitor acquired and digitized multi-parameter physiological waveform data, processed the signals to derive time series (trends) of clinical measures such as heart rate, blood pressures, and oxygen saturation, etc., and also produced bedside monitor alarms. The waveforms (such as electrocardiogram, blood pressures, pulse plethysmograms, respirations) were sampled at 125 Hz, and trend data were updated each minute. The data were subsequently stored temporarily in a central database server that typically supported several ICUs. A customized archiving agent created and stored permanent copies of the physiological data. The data were physically transported from the hospital to the laboratory every 2–4 weeks where they were de-identified, converted to an open source data format, and incorporated into the MIMIC II waveform database. Unfortunately, limited capacity and intermittent failures of the archiving agents limited waveform collection to a fraction of the monitored ICU beds.
2.3 Death Data
The Social Security Death Master files were used to document subsequent dates of death for patients who were discharged alive from the hospital. Such data are important for 28-day and 1-year mortality studies.
3 Data Merger and Organization
A major effort was required in order to organize the diverse collected data into a well-documented relational database containing integrated medical records for each patient. Across the hospital’s clinical databases, patients are identified by their unique Medical Record Numbers and their Fiscal Numbers (the latter uniquely identifies a particular hospitalization for patients who might have been admitted multiple times), which allowed us to merge information from many different hospital sources. The data were finally organized into a comprehensive relational database. More information on database merger, in particular, how database integrity was ensured, is available at the MIMIC-II web site [1]. The database user guide is also online [2].
An additional task was to convert the patient waveform data from Philips’ proprietary format into an open-source format. With assistance from the medical equipment vendor, the waveforms, trends, and alarms were translated into WFDB, an open data format that is used for publicly available databases on the National Institutes of Health-sponsored PhysioNet web site [3].
All data that were integrated into the MIMIC-II database were de-identified in compliance with Health Insurance Portability and Accountability Act standards to facilitate public access to MIMIC-II. Deletion of protected health information from structured data sources was straightforward (e.g., database fields that provide the patient name, date of birth, etc.). We also removed protected health information from the discharge summaries, diagnostic reports, and the approximately 700,000 free-text nursing and respiratory notes in MIMIC-II using an automated algorithm that has been shown to have superior performance in comparison to clinicians in detecting protected health information [4]. This algorithm accommodates the broad spectrum of writing styles in our data set, including personal variations in syntax, abbreviations, and spelling. We have posted the algorithm in open-source form as a general tool to be used by others for de-identification of free-text notes [5].
4 Data Sharing
MIMIC-II is an unprecedented and innovative open research resource that grants researchers from around the world free access to highly granular ICU data and in the process substantially accelerates knowledge creation in the field of critical care medicine. The MIMIC Waveform Database is freely available to all via the PhysioNet website, and no registration is required. The MIMIC Clinical Database is also available without cost. To restrict users to legitimate medical researchers, access to the clinical database requires completion of a simple data use agreement (DUA) and proof that the researcher has completed human subjects training [6].
The MIMIC-II clinical database is available in two forms. In the first form, interested researchers can obtain a flat-file text version of the clinical database and the associated database schema that enables them to reconstruct the database using a database management system of their choice. In the second form, interested researchers can gain limited access to the database through QueryBuilder, a password-protected web service. Database searches using QueryBuilder allow users to familiarize themselves with the database tables and to program database queries using the Structured Query Language. Query output, however, is limited to 1000 rows because of our laboratory’s limited computational resources. Accessing and processing data from MIMIC-II is complex. It is recommended that studies based on the MIMIC-II clinical database be conducted as collaborative efforts that include clinical, statistical, and relational database expertise. Detailed documentation and procedures for obtaining access to MIMIC-II are available at the MIMIC-II web site [1]. The current release of MIMIC-II is version 2.6, containing approximately 36,000 patients, including approximately 7000 neonates, and covering the period 2001–2008. At the present time approximately 1700 individuals worldwide in academia, industry, and medicine have been credentialed to access MIMIC-II and are producing research results in physiologic signal processing, clinical decision support, predictive algorithms in critical care, pharmacovigilance, natural language processing, and more.
5 Updating
In 2008 the hospital made a major change in the ICU information system technology and in ICU documentation procedures. The Philips CareVue system was replaced with iMDsoft’s MetaVision technology. In 2013 we began a major update to MIMIC to incorporate adult ICU data for the period 2008–2012. The effort required learning the entirely new data schema of MetaVision, and merging the new data format with the existing MIMIC design. The new MetaVision data included new data elements such as physician progress notes, oral and bolus medication administration records, etc. Updated data were extracted from hospital archives and from the SSA death files for the newly added patients. Almost two years of effort was invested to acquire, organize, debug, normalize and document the new database before releasing it. MIMIC-III includes 20,000 new adult ICU admissions, bringing the total to approximately 60,000. The new database is known as MIMIC-III, and the acronym has been recast as “Medical Information Mart for Intensive Care” [7].
6 Support
Support of the MIMIC databases includes: credentialing new users, administration of the authorized user list (i.e. users who have signed the DUA and have been granted permission to access MIMIC-II), user account creation, password resets and granting/revoking permissions. The servers providing MIMIC-II include authentication, application, database and web servers. All systems must be monitored, maintained, upgraded and backed up; the maintenance burden continues to increase as the number of database users grows. The engineering staff at LCP attempt to answer user queries as needed. Common questions are added to list of frequently asked questions on the MIMIC website and we regularly update our online documentation.
7 Lessons Learned
Building and distributing MIMIC-like databases is challenging, complex, and requires the cooperation and support of a number of individuals and institutions. A list of some of the more important requirements follows (Table 5.1).
8 Future Directions
The MIMIC-III database is a powerful and flexible research resource, but the generalizability of MIMIC-based studies is somewhat limited by the fact that the data are collected from a single institution. Multi-center data would have the advantages of including wider practice variability, and of course a larger number of cases. Data from international institutions would add still greater strength to the database owing to the even larger variations in practice and patient populations.
Our long-term goal is to create a public, multi-center, international data archive for critical care research. We envisage a massive, detailed, high-resolution ICU data archive containing complete medical records from patients around the world. The difficulty of such a project cannot be understated; nevertheless we propose to lay the foundation for such a system by developing a scalable framework that can readily incorporate data from multiple institutions, capable of supporting research on cohorts of critically ill patients from around the world.
References
MIMIC-II Web Site. http://physionet.org/mimic2
MIMIC User Guide. http://physionet.org/mimic2/UserGuide/
WaveForm DataBase Data Format. http://www.physionet.org/physiotools/wfdb.shtml
Neamatullah I, Douglass M, Lehman LH, Reisner A, Villarroel M, Long WJ, Szolovits P, Moody GB, Mark RG, Clifford GD (2008) Automated de-identification of free-text medical records. BMC Med Inform Decis Mak 8:32. doi:10.1186/1472-6947-8-327
Deidentification Software. http://www.physionet.org/physiotools/deid/
Accessing MIMIC. http://www.physionet.org/mimic2/mimic2_access.shtml
MIMIC-III Website. http://mimic.physionet.org/
Acknowledgments
The development and maintenance of the MIMIC and PhysioNet resources have been funded by the National Institute of Biomedical Imaging and Bioengineering (NIBIB) and the National Institute of General Medical Sciences (NIGMS) over the period 2003 to present. Grants R01EB1659, R01EB017205, R01GM104987, and U01EB008577.
Author information
Authors and Affiliations
Rights and permissions
Open Access This chapter is distributed under the terms of the Creative Commons Attribution-NonCommercial 4.0 International License (http://creativecommons.org/licenses/by-nc/4.0/), which permits any noncommercial use, duplication, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, a link is provided to the Creative Commons license and any changes made are indicated.
The images or other third party material in this chapter are included in the work’s Creative Commons license, unless indicated otherwise in the credit line; if such material is not included in the work’s Creative Commons license and the respective action is not permitted by statutory regulation, users will need to obtain permission from the license holder to duplicate, adapt or reproduce the material.
Copyright information
© 2016 The Author(s)
About this chapter
Cite this chapter
Mark, R. (2016). The Story of MIMIC. In: Secondary Analysis of Electronic Health Records. Springer, Cham. https://doi.org/10.1007/978-3-319-43742-2_5
Download citation
DOI: https://doi.org/10.1007/978-3-319-43742-2_5
Published:
Publisher Name: Springer, Cham
Print ISBN: 978-3-319-43740-8
Online ISBN: 978-3-319-43742-2
eBook Packages: MedicineMedicine (R0)