Abstract
Biodiversity is essential to life on Earth and motivates many efforts to collect data about species. These data are collected in different places and published in different formats. Researchers use it to extract new knowledge about living things, but it is difficult to retrieve, combine and integrate data sources from different places. This work will investigate how to integrate biodiversity information from heterogeneous sources using Semantic Web technologies. Its main objective is to propose an architecture to link biodiversity data using mainly their spatiotemporal dimension, effectively search these linked data sets and test them using real use cases, defined with the help of biodiversity experts. It is also an important objective to propose a suitable provenance model that captures not only data origin but also temporal information. This architecture will be tested on a set of representative data from important Brazilian institutions that are involved in studies of biodiversity.
You have full access to this open access chapter, Download conference paper PDF
Similar content being viewed by others
Keywords
1 Problem Statement
Biological diversity is essential to life sustainability on Earth [1]. The large amount of data generated by researchers in biodiversity has led to discussions about how to find the best ways to organize this data and provide tools and environments that stimulate and facilitate the search for information. Currently, when using search tools for biodiversity data, experts specify their queries using one or more terms of interest. However, these terms may not match those that are part of the documents and, therefore, some relevant documents are not recovered [2].
In Brazil, there is a network of Amazonian and extra-Amazonian institutions that are involved in studies of biodiversity. This network is integrated by important institutions, such as the National Research Institute for the Amazon (INPA)Footnote 1, the National Institute for Space Research (INPE)Footnote 2, the Global Biodiversity Information Facility (GBIF)Footnote 3, the Emilio Gueldi Museum in Par? (MPEG)Footnote 4, and Brazilian Agricultural Research Corporation (EMBRAPA)Footnote 5. These organizations collect and contribute large amounts of data about biodiversity. One of the most frequent problems, reported by biodiversity researches, is how to retrieve and integrate information simultaneously from the big number of data sources found on the various biodiversity databases. Typically, these users utilize the biodiversity data to visualize integrated information about the collected specimens [1].
The problem is that a specialist may specify one or more terms (strings) for a search and, due to the large amount of available data, get responses with too many results (not all relevant) [1]. He then has a lot of work sifting through the results for the desired information, because the results provided are very broad and may not even contain the targeted data. This activity is not particularly well supported by biodiversity software tools based on keyword searching (the kind usually found in the Web) [2].
Even if a search is successful, it is the biodiversity specialist who must browse the selected documents to extract the information he/she is looking for. There is not much support for retrieving the actual information from the documents, a very time-consuming activity, and put it in a suitable format [1]. Of course, there are tools that can retrieve texts, split them into parts, check the spelling, and count their words. But, when it comes to interpret sentences and extract useful information for biodiversity specialists, the capabilities of current software are still very limited. It is simply very difficult to distinguish the meaning of the following query:
Return all occurrences of records of insects that belong to the ant family (Formicidae) and have been found in an aquatic habitat in the Brazilian Amazon forest
For instance, an SQL query, in a traditional database, would only succeed if records have the exact information (strings) asked in the query. In this case, a record of a Paraponera clavata specimen (bullet-ant) that was found in a swamp would not be returned. The strings Paraponera clavata and swamp are not in the query.
Biodiversity specialists also need more complex queries, e.g., requiring spatiotemporal query processing, such as deriving co-occurrence of species in a given spacetime frame. Such processing is seldom supported. Other queries involve biodiversity relations among species, e.g., farms within a protected area. Such relationships are not stored, and must be deduced by the scientist after performing a sequence of queries and simulations.
2 Research Questions
The main question research is:
-
How can we integrate biodiversity information from heterogeneous sources using their spatial location and temporal data?
To answer this question, we also need to find an answer to the following questions:
-
How can we improve the interoperability of the biodiversity data?
-
How can we improve the location accuracy of biodiversity data?
-
How to improve the trust in biodiversity data?
3 Hypotheses
The main hypotheses related to this research are:
-
Representing biodiversity data as Linked Data will improve the integration it with data from different and independent data sources (if they share common ontology terms).
-
Using biodiversity data as Linked Data will resolve advanced and complex querying that was not possible before.
-
Capturing the spatiotemporal characteristic from biodiversity data will perform more accurate locations.
-
Reusing the provenance model will improve the trust of the biodiversity datasets and scientists could trust the data links provided by the network of Amazonian and extra-Amazonian institutions.
4 Research Approach
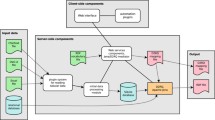
Initially, we will analyze and extract spatiotemporal data of biodiversity and geographic databases (such as soil, rivers, deforestation) from different data sources (INPE, INPA, MPEG, EMBRAPA). Once the spatiotemporal data is extracted, the next step is to find the links between different sources. For this reason, we will identify the vocabularies and ontologies with specific relationships to biodiversity and geospatial information. Following this, we will map biodiversity data and the ontologies describing them, considering data provenance. We will convert biodiversity data in the Semantic Web format (mapping). In order to provide a better feedback on the quality of the data. The mapping will be implemented using state of the art Semantic Web tools and tested on a set of representative data about biodiversity.
We will then develop a new Linked Data architecture to integrate biodiversity information from heterogeneous sources using their spatial location and temporal data. A first prototype, based on this architecture, will be implemented. This prototype will permit data integration from different triple stores, checks for inconsistencies and new knowledge extraction. The generated linked information will be retrievable in a friendly way. After that, an experimentation phase, based on controlled experiments, will be carried out. To conclude, we will test various use cases.
5 Evaluation Plan
There are different aspects of the proposed architecture which need to be assessed:
-
The interlinking between biodiversity vocabularies and ontologies with other domains. Interlinking is provided by RDF triples that establish a link between the entity identified by the subject with the entity identified by the object.
-
The performance in process complexity SPARQL and GeoSparql queries.
-
The accuracy, precision and recall of the retrieved links in conjunction with other domains.
6 Related Work
In this Section, we will review the related work on the use of Linked Data and Provenance in biodiversity domain.
Linked Data is gaining traction in the scientific community. One of the earliest investigation relates with Amazon Rainforest was conducted by Cardoso et al. [3]. They describe a geographical gazetteer that associates place names to geographic coordinate data from two large biodiversity repositories: GBIF and the SpeciesLinkFootnote 6. However, there is still a fundamental lack to answer complex queries with spatiotemporal characteristics (e.g., farms within a protected area between 2005 and 2011).
Kauppinen et al. [4] describe the Linked Brazilian Amazon Rainforest Dataset (LBARD) using ontologies and vocabularies. However, the authors only show the Amazon Rainforest data using the R program. Users have to invest a considerable amount of time in programming in R, and perform many manual tasks, to obtain the needed datasets.
Garcia et al. [5] propose a data mining framework for primary biodiversity data analysis. This approach uses relational database to store the biodiversity data. Rocca-Serra et al. [6] describe how resources of the Open Biological and Biomedical Ontologies (OBO)Footnote 7 have been used to provide a semantic framework enabling the presentation of biodiversity information as Linked Data. Wieczorek et al. [7] describe the Darwin Core data standard for publishing and integrating biodiversity information. We plan to use the Darwin Core standard to capture complex aspects of the biodiversity domain.
A critical look at the available literature indicates that most of existing approaches suffer of the following limitations: (i) A number of techniques have been developed for using ontologies to retrieve relevant documents in response to a query. However, none of the works focused on the problem of storage, retrieval and link RDF triples using their spatiotemporal information. (ii) The approaches do not provide an explicit visualization of the geospatial and biodiversity dataset. There is still a fundamental lack of approaches to visualizing linked biodiversity data that use spatial and temporal relations.
Provenance describes how a data object came to be in its present state, and thus, it describes the evolution of the object over time [8]. There are a number of studies, which have used provenance in the biodiveristy domain [9–11]. For example, Beserra et al. [11] propose a provenance-based approach to manage long term preservation of scientific data. Their approach is based on the Open Provenance Model (OPM) [12]. However, this approach does not provide support to connect curated metadata with LOD, which would allow breaking down disciplinary boundaries among repositories and enhance reuse.
The PROV specificationFootnote 8 defines a core data model for provenance for building representations of the entities, people and processes involved in producing a piece of data or thing in the world. However, there is a lack of expressiveness using this generic W3C recommendation to model the different types of organisms that co-occur in time and space (geospatial relations).
A critical look at the available literature indicates that a number of techniques have been developed for using provenance models, such as OPM and DCMI, in the different scientific domains. Despite the variety of models, there is currently no unified, conceptual model for biodiversity information and provenance that can be applied to different datasets and setups, while remaining both expressive and generic enough to cover many use cases.
7 Reflections
The main difference of this thesis proposal compared to existing works on linked biodiversity data is that we (i) introduce the idea of use the spatiotemporal information from biodiversity heterogeneous sources data to interlinking with other domains; and (ii) another important facet, when dealing with scientific data, is provenance. We plan to specialize the PROV provenance model for biodiversity data.
References
Magnusson, W., Braga-Neto, R., Pezzini, F., Baccaro, F., Bergallo, H., Penha, J., de Jesus Rodrigues, D., Verdade, L.M., Lima, A., Albernaz, A.L., Hero, J.M., Lawson, B., Castilho, C., Drucker, D., Franklin, E., Medonca, F., Costa, F., Galdino, G., Castley, G., Zuanon, J., do Vale, J., dos Santos, J.L.C., Luizao, R., Cintra, R., Barbosa, R.I., Lisboa, A., Koblitz, R., da Cunha, C.N., Pontes, A.R.M.: Biodiversity and Integrated Environmental Monitoring. Program for Planned Biodiversity and Ecosystem Research (PPBio) (2013)
Amanqui, F.K., Serique, K.J., Cardoso, S.D., Santos, J.L., Albuquerque, A., Moreira, D.A.: Improving biodiversity data retrieval through semantic search and ontologies. In: 2014 IEEE/WIC/ACM International Joint Conferences on Web Intelligence (WI) and Intelligent Agent Technologies (IAT), vol. 1, pp. 274–281, August 2014
Cardoso, S.D., Amanqui, F.K., Serique, K.J., dos Santos, J.L., Moreira, D.A.: SWI: a semantic web interactive gazetteer to support linked open data. Future Gener. Comput. Syst. 54, 389–398 (2015)
Kauppinen, T., de Espindola, G.M., Jones, J., Sanchez, A., Gräler, B., Bartoschek, T.: Linked brazilian amazon rainforest data. Semant. Web J. 5(2), 151–155 (2014)
Fontes, S.G., Stanzani, S.L., Correa, P.L.P.: A data mining framework for primary biodiversity data analysis. In: Rocha, A., Correia, A.M., Costanzo, S., Reis, L.P. (eds.) New Contributions in Information Systems and Technologies: Volume 1. AISC, vol. 353, pp. 813–821. Springer, Heidelberg (2015)
Rocca-Serra, P., Walls, R., Parnell, J., Gallery, R., Zheng, J., Sansone, S.A., Gonzalez-Beltran, A.: Modeling a microbial community and biodiversity assay with OBO foundry ontologies: the interoperability gains of a modular approach. Database 2015 (2015)
Wieczorek, J., Bloom, D., Guralnick, R., Blum, S., Döring, M., Giovanni, R., Robertson, T., Vieglais, D.: Darwin core: an evolving community-developed biodiversity data standard. PLoS ONE 7(1), 1–8 (2012)
Omitola, T., Gibbins, N., Shadbolt, N.: Provenance in linked data integration (2010)
Zhao, J., Klyne, G., Shotton, D.: Provenance and linked data in biological data webs. In: Bizer, C., Heath, T., Idehen, K., Berners-Lee, T. (eds.) Proceedings of the WWW 2008 Workshop on Linked Data on the Web (LDOW 2008) (2008)
Wang, S., Padmanabhan, A., Myers, J.D., Tang, W., Liu, Y.: Towards provenance aware geographic information systems. In: Proceedings of the 16th ACM SIGSPATIAL International Conference on Advances in Geographic Information Systems, GIS 2008, p. 70:170:4. ACM, New York (2008)
Beserra Sousa, R., Cintra Cugler, D., Gonzales Malaverri, J., Bauzer Medeiros, C.: A provenance-based approach to manage long term preservation of scientic data. In: 2014 IEEE 30th International Conference on Data Engineering Workshops (ICDEW), pp. 162–133, March 2014
Moreau, L., Freire, J., Futrelle, J., McGrath, R.E., Myers, J., Paulson, P.: The open provenance model: an overview. In: Freire, J., Koop, D., Moreau, L. (eds.) IPAW 2008. LNCS, vol. 5272, pp. 323–326. Springer, Heidelberg (2008)
Acknowledgments
The research activities described in this paper were funded by Ghent University, iMinds, the IWT-Flanders, the FWO-Flanders, and the European Union, and the FINCyT Science and Technology Program from Peru.
Author information
Authors and Affiliations
Corresponding author
Editor information
Editors and Affiliations
Rights and permissions
Copyright information
© 2016 Springer International Publishing Switzerland
About this paper
Cite this paper
Amanqui, F., Verborgh, R., Mannens, E., Van de Walle, R., Moreira, D. (2016). Using Spatiotemporal Information to Integrate Heterogeneous Biodiversity Semantic Data. In: Bozzon, A., Cudre-Maroux, P., Pautasso, C. (eds) Web Engineering. ICWE 2016. Lecture Notes in Computer Science(), vol 9671. Springer, Cham. https://doi.org/10.1007/978-3-319-38791-8_41
Download citation
DOI: https://doi.org/10.1007/978-3-319-38791-8_41
Published:
Publisher Name: Springer, Cham
Print ISBN: 978-3-319-38790-1
Online ISBN: 978-3-319-38791-8
eBook Packages: Computer ScienceComputer Science (R0)